
Contact
Email: osnat@umd.edu
Call: (240) 314-6245
Osnat Herzberg
Professor
Herzberg Group
Contact
Email: osnat@umd.edu
Call: (240) 314-6245
Education
- Ph.D., Chemistry, Weizmann Institute of Science, Israel, 1982
- M.S., Chemistry, Weizmann Institute of Science, Israel, 1976
- B.S., Chemistry, Technion, Israel, 1971
Profile
Dr. Osnat Herzberg is a structural biologist interested in the relationship between the function and structure of proteins and how protein-ligand interactions can guide drug discovery. The Herzberg lab uses X-ray crystallography and other biophysical, biochemical, and cellular approaches to better understand various experimental systems.
CURRENT RESEARCH
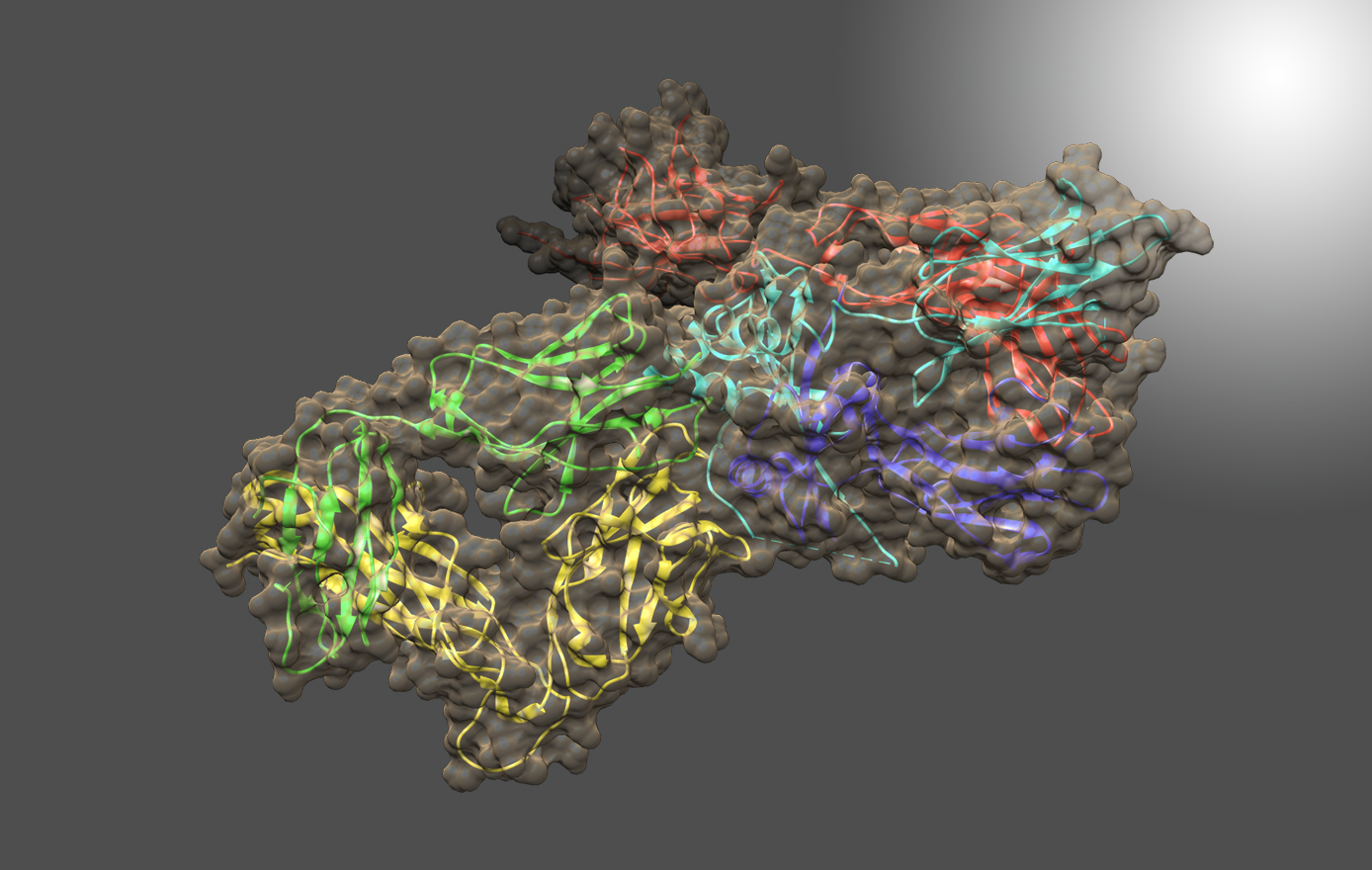
Current projects in the Herzberg lab include studies of genetic factors that contribute to inflammatory bowel diseases; understanding the structure and function of myovirus phage CBA120 proteins, a bacteriophage that kills pathogenic E. coli; and, drug discovery for combating the parasitic diseases giardiasis and amebiasis.
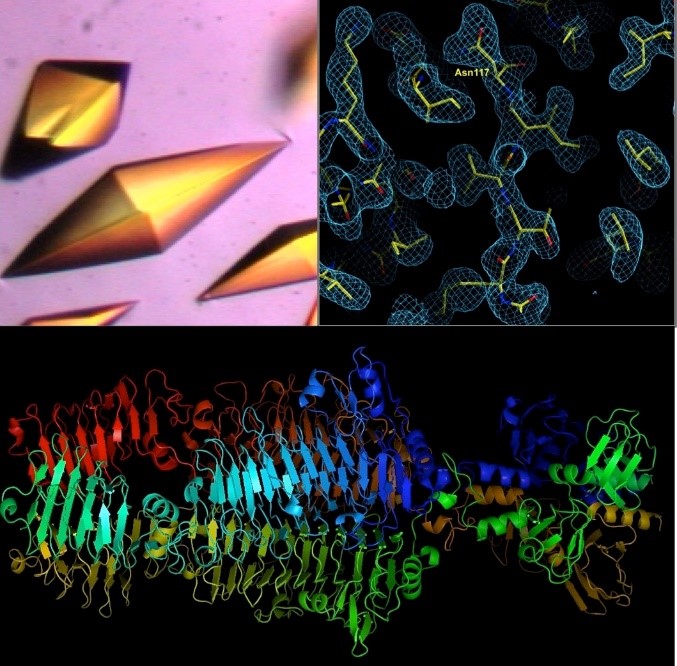
A tailspike protein structure is shown at the bottom.
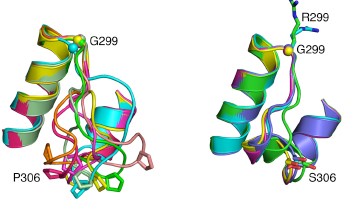
The inflammatory bowel diseases (IBD), Crohn’s disease and ulcerative colitis, are human complex trait diseases caused by multiple genetic variants and environmental factors. The Herzberg lab is working to identify genetic variants known as single nucleotide polymorphisms (SNPs) that are associated with these diseases. They hope to determine how these variants alter proteins that contribute to disease. This is a collaborative project involving Dr. John Moult’s group. Dr. Moult’s group is identifying candidate disease variants by computational methods and the Herzberg group is carrying out experimental studies to validate these findings. One particular protein, called gasdermin B, has variants that have been implicated in IBD. Studies by the Herzberg lab revealed that protein changes associated with different SNPs alter the flexibility of a protein loop region and these changes have the potential to alter protein-protein interactions. Dr. Herzberg’s lab also discovered that gasdermin B can bind to lipid molecules that are critical to maintaining the integrity of the gut wall. It remains to be determined whether and how the gasdermin B variant found in IBD patients affects the lipid composition of the gut wall.
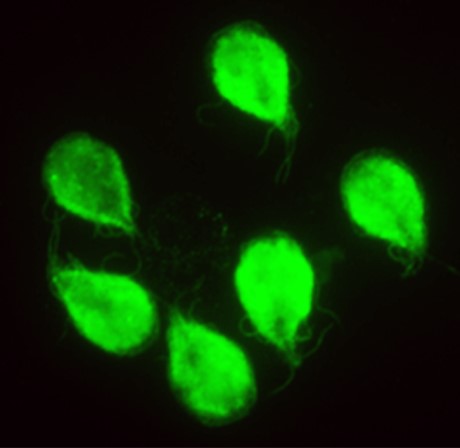
The parasites Giardia lamblia and Entamoeba histolytica cause severe intestinal diseases and are major health hazards in undeveloped and developing countries. Moreover, disease outbreaks in the developed world are associated with drug resistance, which is rapidly spreading, and many patients do not tolerate these standard-of-care drugs. The Herzberg lab is developing less toxic drugs that are not prone to drug resistance. The Herzberg group is optimizing the properties of a drug called fumagillin, which is used in the European Union to treat immune compromised patients suffering from the parasitic infection microsporidiosis, in order to develop new and better antigiardiasis and antiamebiasis therapeutics. Compound optimization is facilitated by the unique capabilities of the group that integrate all aspects of preclinical studies, including medicinal chemistry, biochemistry, molecular biology, cellular biology, and animal studies.