New Term ‘Translon’ Proposed to Standardize Genetic Technology
Tue, Oct 28, 2025
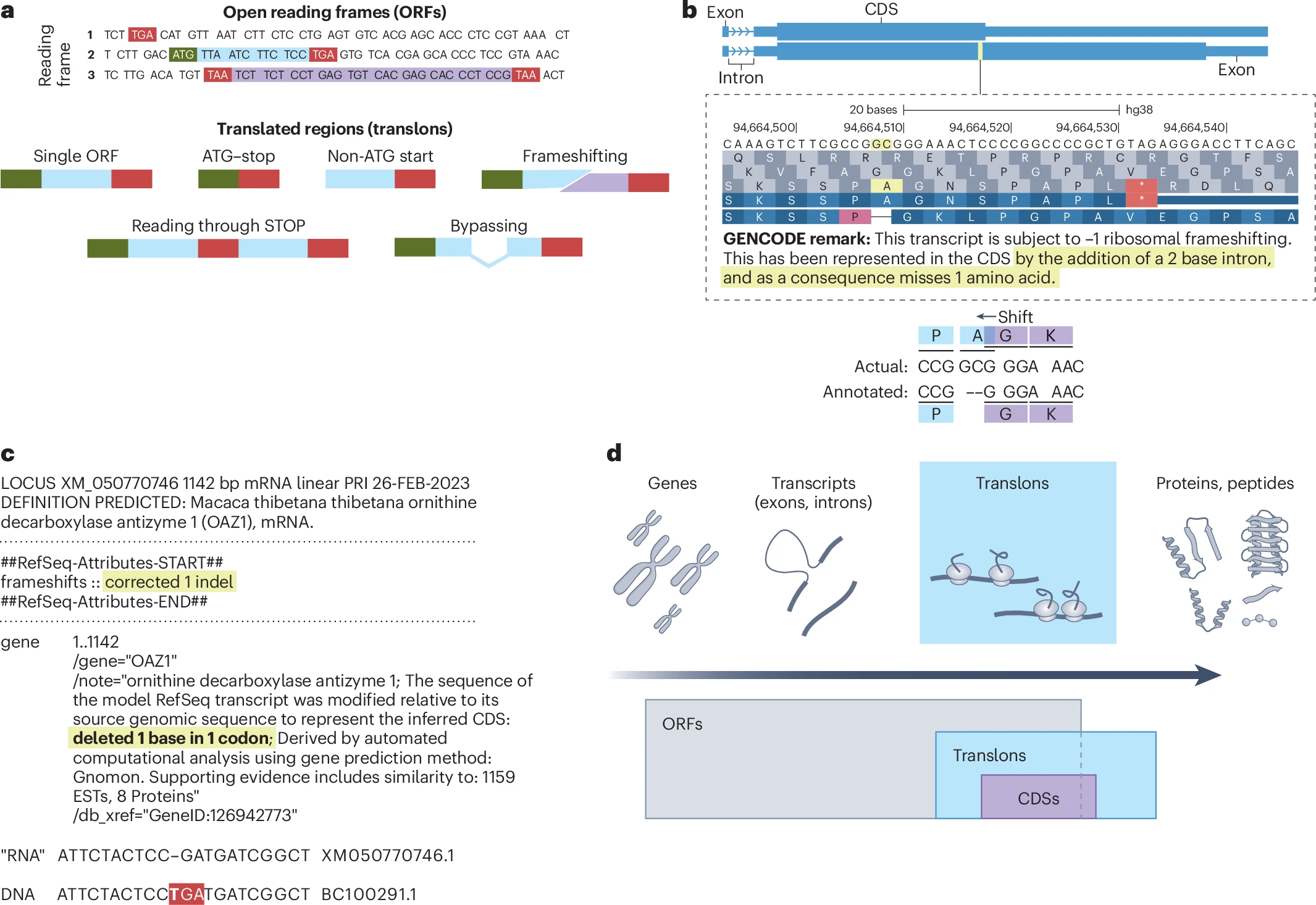
A recent publication in Nature Methods introduced a new term, ‘translon,’ to denote any region of genetic code that is translated. The article, entitled "Translon: a single term for translated regions,” was co-authored by an international team of researchers including Dr. Jonathan Dinman, Director of IBBR.
In molecular biology, translation is the process where cells use RNA as a template to build proteins, which are essential for nearly every function in the body. Ribosomes, tiny molecular machines, carry out this translation, reading the RNA sequence and assembling proteins accordingly.
Currently, a large variety of terms are being used such as “open reading frames” (ORFs), “small ORFs” (smORFs), and “non-canonical ORFs” (ncORF). These terms often overlap with or conflict with each other, and sometimes even contradict, leading to a lack of consistency and clarity. These terms focus on how the sequence is structured or what it might produce, rather than the simple fact that it is being read and translated by the ribosome.
By introducing the term ‘translon,’ researchers aim to clear up some of the confusion with a single, universal term for any sequence read by a ribosome, regardless of its length, structure, or functionality. This term will be especially useful as studies increasingly discover new, previously unrecognized regions being translated in cells.
Having a clear, consistent term enables researchers to more accurately describe and study gene expression. This effort also closely aligns with NIST’s broader mission of standardization and scientific consistency, whether it's in regard to measurements, weights, or terminology.
To read the full article, click here.