Contact
Email: grishaev@umd.edu
Call: (240) 314-6892
Alexander Grishaev
Grishaev Group
Contact
Email: grishaev@umd.edu
Call: (240) 314-6892
Education
- Postdoctoral Fellow, Laboratory of Chemical Physics, NIDDK/NIH, 2001-2005
- Ph.D., Chemistry/Biophysics, Carnegie Mellon University, 2001
- M.S., Chemistry, University of Wisconsin-Madison, 1995
- Diploma, Physical/Quantum Chemistry, Moscow State University, 1993
Profile
Dr. Alexander Grishaev’s laboratory focuses on integrative structural biology and deriving biomolecular structures by combining experimental data from several complementary, biophysical techniques within a computational framework that optimally restrains the conformation space. The lab’s primary experimental approaches are solution X-ray and neutron scattering, and nuclear magnetic resonance (NMR) spectroscopy. They aim to design methodologies for maximizing the information content and fidelity of interpretation of the observables attainable by these techniques. An important part of the Grishaev lab’s work is mining information in structural databases to improve force fields for protein/RNA/DNA structure refinement. The group focuses on systems characterized by a low density of experimental restraints, such as flexibly linked multi-domain or disordered proteins, amyloid fibrils, protein complexes, and RNA constructs.
CURRENT RESEARCH
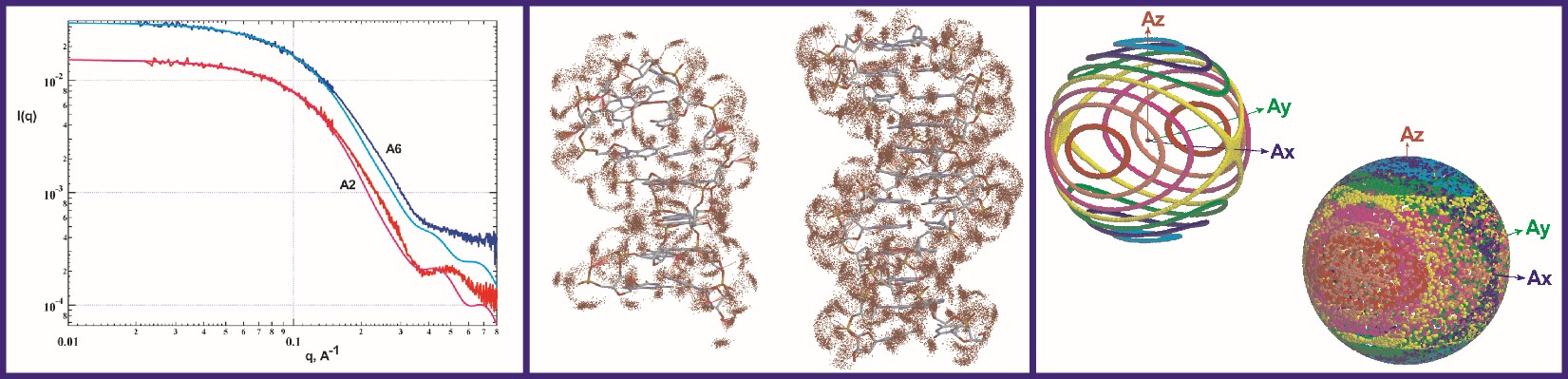
Using wide-angle solution X-ray scattering to define biomolecular structure and dynamics

The Grishaev lab develops methods for improving both the speed and the accuracy of modeling solution X-ray scattering data from atomic coordinates to facilitate their integration into structure refinement of intrinsically disordered and flexibly linked multi-domain proteins, RNA, DNA, and lipid or detergent-based nanoparticles. The lab’s ultimate goal is to determine the structures of proteins and RNA as multi-state conformational ensembles, which they view as the next frontier of structural biology. Model improvements are underpinned by high-fidelity molecular dynamics simulations in explicit solvent and structural database analysis, allowing formulation of detailed, experimentally verifiable, multi-dimensional hydration maps and motional models of biomolecules. Infrastructure support for this work comes from state-of-the-art facilities including in-house small- and wide-angle X-ray scattering (SAXS/WAXS) instrumentation, synchrotron beamlines for both non-resonant and anomalous scattering experiments, local neutron scattering facilities at NIST, and a high-performance computing cluster at IBBR. The Grishaev group’s strategy of jointly using wide-angle solution scattering data, NMR, and advanced simulation techniques aims to maximize the density of the experimental observables while limiting the expansion of the number of degrees of freedom to minimize data over-fitting. The group also develops novel scattering approaches -- including high-contrast heavy atoms labels and anisotropic alignment -- to increase the information content of scattering data.
Improving the accuracy of NMR-determined biomolecular structures via better force fields and advanced simulation techniques
NMR is an important component of the lab’s workflow and their efforts include increasing the impact of NMR restraints, such as residual dipolar couplings and chemical shifts. They develop both novel empirical energy terms and codes for better use of NMR and SAXS data, with applications to structure refinement packages, such as Xplor-NIH, CNS, and Amber. They also develop and apply metrics for both empirical protein and RNA structure validation and experimental cross-validation as sensitive and objective gauges of the coordinates’ accuracy.
.