
Brian Pierce
Associate Professor
Pierce Group (240) 314-6271 pierce@umd.eduDr. Brian Pierce’s laboratory develops and applies computer algorithms to better understand how the immune system recognizes pathogens and cancer, and his lab is particularly interested in antibodies, T cell receptors, and vaccine design. For more information on the Pierce Lab, visit the lab web site at: https://piercelab.ibbr.umd.edu.
CURRENT RESEARCH
Recent efforts have focused on studying the structure of the hepatitis C virus (HCV) to inform the design of novel vaccine candidates. This work includes the development of a pioneering epitope-based vaccine for HCV that elicits neutralizing antibodies. The Pierce lab is also modeling how HCV can escape antibody neutralization, and this can be used toward developing new vaccine candidates.
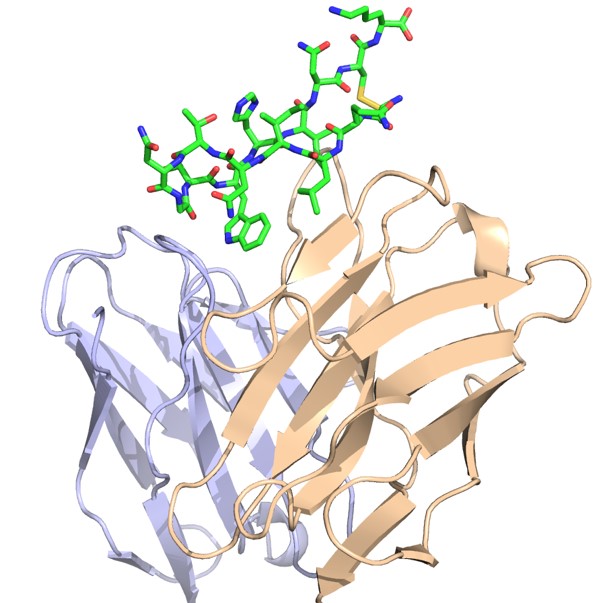
The Pierce group is working on understanding and predicting how antibodies recognize viruses and other pathogens. Through a collaboration with researchers at Stanford University, they now have an unprecedented view of the structure and key antibody recognition features of HCV. This provides a roadmap for improved HCV vaccine candidates, as well as insights into antibody recognition of viral envelope proteins in general.
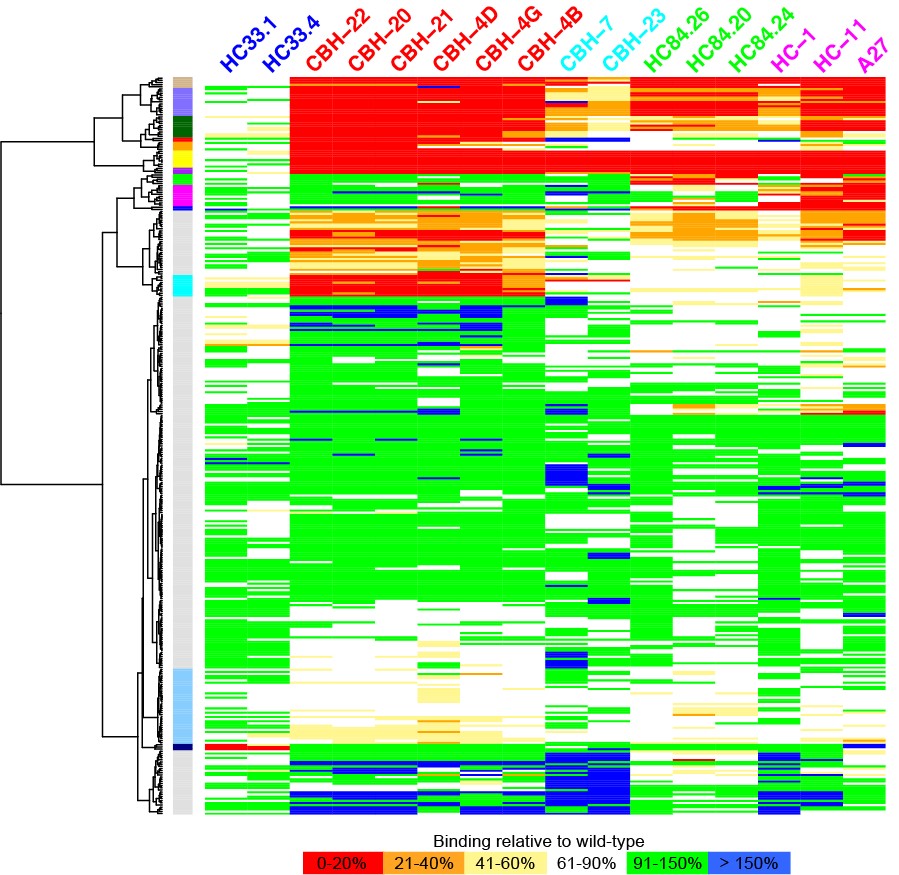
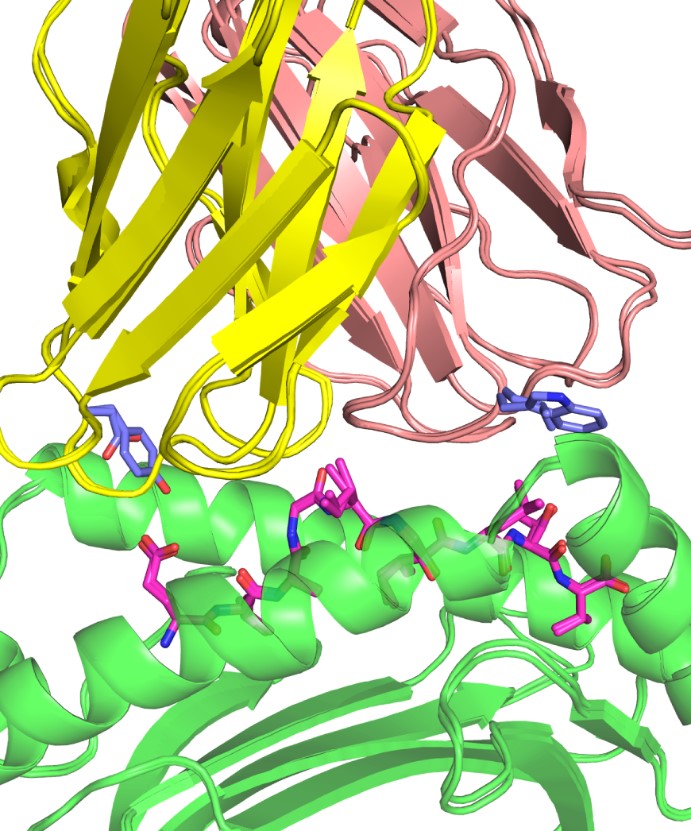
A longstanding area of focus in the lab has been understanding how T cells recognize specific antigens, and work in the Pierce lab includes modeling how T cell receptors (TCR) can be engineered to target cancer cells. The group has developed a therapeutic TCR that targets melanoma, and ongoing work includes developing algorithms to improve and optimize TCR structures for immunotherapeutic applications.
The Pierce lab has developed a number of predictive protein modeling and design algorithms to carry out their research, including Rosetta, TCRFlexDock, RosettaTCR, ZRANK, and ZDOCK. The lab is a member of the RosettaCommons community, which is a global network of developers of the Rosetta modeling and design software.
Publications
- Structural insights into clonal restriction and diversity in T cell recognition of two immunodominant SARS-CoV-2 nucleocapsid epitopes.
- Dynamic S-acylation of GSDMA regulates pyroptosis.
- Evaluation of Alphafold modeling for elucidation of nanobody-peptide epitope interactions.
- Computational Prioritization of T Cell Epitopes to Overcome HLA Restriction and Antigenic Diversity in Plasmodium falciparum.
- Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
- Evaluation of Alphafold modeling for elucidation of nanobody-peptide epitope interactions.
- A comprehensive engineering strategy improves potency and manufacturability of a near pan-neutralizing antibody against HIV.
- RNA Helicase DDX3 Interacts with the Capsid Protein of Hepatitis E Virus and Plays a Vital Role in the Viral Replication.
- AlphaFold and Docking Approaches for Antibody-Antigen and Other Targets: Insights From CAPRI Rounds 47-55.
- Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
- Exploring the potential of structure-based deep learning approaches for T cell receptor design.
- TCR3d 2.0: expanding the T cell receptor structure database with new structures, tools and interactions.
- Hepatitis C Virus E1E2 Structure, Diversity, and Implications for Vaccine Development.
- Proscan: a structure-based proline design web server.
- Combinatorially restricted computational design of protein-protein interfaces to produce IgG heterodimers.
- A single C-terminal residue controls SARS-CoV-2 spike trafficking and incorporation into VLPs.
- Evaluation of AlphaFold Antibody-Antigen Modeling with Implications for Improving Predictive Accuracy.
- Structural basis for T cell recognition of cancer neoantigens and implications for predicting neoepitope immunogenicity.
- Impact of AlphaFold on structure prediction of protein complexes: The CASP15-CAPRI experiment.
- SARS-CoV-2 infection establishes a stable and age-independent CD8+ T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
- Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
- TCRmodel2: high-resolution modeling of T cell receptor recognition using deep learning.
- Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
- Mucosal nanobody IgA as inhalable and affordable prophylactic and therapeutic treatment against SARS-CoV-2 and emerging variants.
- Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants.
- Structural Features of Antibody-Peptide Recognition.
- Induction of broadly neutralizing antibodies using a secreted form of the hepatitis C virus E1E2 heterodimer as a vaccine candidate.
- An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking.
- T cell receptors (TCRs) employ diverse strategies to target a p53 cancer neoantigen.
- Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
- Molecular Determinants of Filament Capping Proteins Required for the Formation of Functional Flagella in Gram-Negative Bacteria.
- An Antigenically Diverse, Representative Panel of Envelope Glycoproteins for Hepatitis C Virus Vaccine Development.
- Structural and energetic profiling of SARS-CoV-2 receptor binding domain antibody recognition and the impact of circulating variants.
- Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment.
- T Cell Receptor Genotype and Ubash3a Determine Susceptibility to Rat Autoimmune Diabetes.
- Structural and Biophysical Characterization of the HCV E1E2 Heterodimer for Vaccine Development.
- Structure-Based and Rational Design of a Hepatitis C Virus Vaccine.
- An expanded benchmark for antibody-antigen docking and affinity prediction reveals insights into antibody recognition determinants.
- Anti-CfaE nanobodies provide broad cross-protection against major pathogenic enterotoxigenic Escherichia coli strains, with implications for vaccine design.
- Design of a native-like secreted form of the hepatitis C virus E1E2 heterodimer.
- High-throughput modeling and scoring of TCR-pMHC complexes to predict cross-reactive peptides.
- CoV3D: a database of high resolution coronavirus protein structures.
- Structure-Based Design of Hepatitis C Virus E2 Glycoprotein Improves Serum Binding and Cross-Neutralization.
- CoV3D: A database and resource for high resolution coronavirus protein structures.
- In Vivo and In Vitro Potency of Polyphosphazene Immunoadjuvants with Hepatitis C Virus Antigen and the Role of Their Supramolecular Assembly.
- Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
- Macromolecular modeling and design in Rosetta: recent methods and frameworks.
- Cellular origins and genetic landscape of cutaneous gamma delta T cell lymphomas.
- Modeling and Viewing T Cell Receptors Using TCRmodel and TCR3d.
- Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment.
- Geometrical characterization of T cell receptor binding modes reveals class-specific binding to maximize access to antigen.
- Newcastle disease virus vectors expressing consensus sequence of the H7 HA protein protect broiler chickens and turkeys against highly pathogenic H7N8 virus.
- TCR3d: The T cell receptor structural repertoire database.
- Broadly neutralizing antibodies from an individual that naturally cleared multiple hepatitis C virus infections uncover molecular determinants for E2 targeting and vaccine design.
- Antigenicity and Immunogenicity of Differentially Glycosylated Hepatitis C Virus E2 Envelope Proteins Expressed in Mammalian and Insect Cells.
- Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation.
- Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites.
- Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine.
- Computational Modeling of Hepatitis C Virus Envelope Glycoprotein Structure and Recognition.
- TCRmodel: high resolution modeling of T cell receptors from sequence.
- Designing a B Cell-Based Vaccine against a Highly Variable Hepatitis C Virus.
- Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design.
- Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice.
- Viral evasion and challenges of hepatitis C virus vaccine development.
- A generalized framework for computational design and mutational scanning of T-cell receptor binding interfaces.
- Hepatitis C virus vaccine candidates inducing protective neutralizing antibodies.
- Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment.
- Computational Modeling of T Cell Receptor Complexes.
- Computational Reprogramming of T Cell Antigen Receptor Binding Properties.
- How structural adaptability exists alongside HLA-A2 bias in the human αβ TCR repertoire.
