
Curtis Meuse
Research Chemist
Meuse Group (240) 314-6131 curtis.meuse@nist.govDr. Curt Meuse’s research focuses on the development of accurate and precise techniques for the characterization of higher order structure of biopharmaceuticals.
Biopharmaceuticals represent a valuable and important sector of the US economy. Advanced methods are required to quantify the molecular composition and conformation of industrially relevant protein products such as protein biopharmaceuticals, protein arrays, membrane proteins for research purposes, and protein standards. Such measurements allow comparisons of protocols and determination of mechanisms of action. Dr. Meuse’s work focuses on the development and standardization of methods to characterize biologically active states, to measure structural changes, and to explore physical processes such as aggregation and binding that contribute to biological inactivation of proteins. For example, he has developed infrared spectroscopic methods to quantify protein structural stability and protein binding interactions using deuterium exchange; that is, by quantifying the extent of the exchange of deuterium for hydrogen in the amide bonds of proteins in solutions, as solids, in membranes, or immobilized on surfaces. In addition, collaborations with the United Kingdom’s National Physics Laboratory led to a pilot study for the standardization of biomolecular circular dichroism measurements under the auspices of the International Committee for Weights and Measures.
CURRENT RESEARCH
Characterizing Sub-populations of Protein Biologics using Fourier Transform Infrared Spectroscopy (FT-IR)
“…current analytical methods fall short in assessing the higher-order structure of biopharmaceuticals, i.e. in detecting the presence of small, conformationally altered populations… in a dynamic equilibrium...” (Berkowitz et al. 2012. Nature Rev. Drug Discov)

1519 tryp < 1636 β-sheet < 1654 α-helix < 1665 Hinge loop < 1613 Asp (NH2) < 1678 Asp (C=O) < 1541 Glu < 1559 Glu < 1572 Asp < 1693 β-turn < 1727 Phe/Try (in collaboration with Protein Dynamic Solutions, LLC)
Dr. Meuse’s work suggests that methods for automated sampling and sample manipulation during measurements, and the application of correlation analyses to these robust data sets, are key to developing techniques for characterizing subpopulations of proteins with different tertiary and quaternary structures to aid in biopharmaceutical development. To this end, he explores methods of submultiple data analysis (below), Attenuated Total Reflection (ATR), sample buffer modulation, and microscopic 2-D analysis of protein temperature profiles (right).
Submultiple Data Collection to Improve Spectroscopy
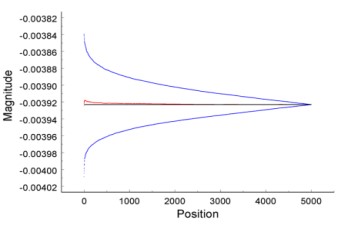
(Meuse et al. 2018. Anal. Chem.)
By averaging over time, the noise on a spectrometric signal can be reduced, and thus the signal-to-noise ratio of the spectrum can be increased and sample bias reduced. The averaged noise is expected to decrease as the square root of the data collection time (t½). However, with contemporary capabilities, during the same collection time, the signal can be averaged over much shorter, equal, fixed periods. This is the set of signals over submultiples of the total collection time. With a sufficiently large set of submultiples, the distribution of the signal's fluctuations over the submultiple periods of the data stream can be acquired at each wavelength (or frequency). From the autocorrelations of submultiple sets, the fractions of stochastic and non-stochastic fluctuations can be determined. Some of the fluctuations are fast drift, that is, drift over a time shorter than the complete measurement period of the average spectrum. What is usually assumed to be stochastic noise has a significant component of fast drift due to changes of conditions in the spectroscopic system. With this new perspective, we suggest that automated and/or correlated measurement methods will help remove fast drift from biopharmaceutical measurements of subpopulations.
Publications
- Quantitative infrared spectroscopy of formalin-fixed, paraffin-embedded tissue specimens: paraffin wax removal with organic solvents.
- Mechanism of formation of vesicle fused phospholipid monolayers on alkanethiol self-assembled monolayer supports.
- Tertiary structure changes in albumin upon surface adsorption observed via fourier transform infrared spectroscopy.
- Infrared techniques for quantifying protein structural stability.
- Molecular dynamics study of surface-tethered S(CH2CH2O)6CH3: helix formation and thermal disorder.
- Electrogenic proton-pumping capabilities of the m-fast and m-slow photocycles of bacteriorhodopsin.
- The ability of actinic light to modify the bacteriorhodopsin photocycle revisited: heterogeneity vs photocooperativity.
- Detection of operons.
- SNPs3D: candidate gene and SNP selection for association studies.
- Rigorous performance evaluation in protein structure modelling and implications for computational biology.
- Osteoblast cell membrane hybrid bilayers for studying cell-cell interactions.
- Towards computing with proteins.
- Identification and analysis of deleterious human SNPs.
- Critical assessment of methods of protein structure prediction (CASP)--round 6.
- Progress over the first decade of CASP experiments.
- Protein family clustering for structural genomics.
- Loss of protein structure stability as a major causative factor in monogenic disease.
- The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses.
- A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction.
- Isostructural self-assembled monolayers. 2. Methyl 1-(3-mercaptopropyl)-oligo(ethylene oxide)s.
- Critical assessment of methods of protein structure prediction (CASP)-round V.
- Three-dimensional structural location and molecular functional effects of missense SNPs in the T cell receptor Vbeta domain.
- Assessment of progress over the CASP experiments.
- Evaluation of disorder predictions in CASP5.
- CAPRI: a Critical Assessment of PRedicted Interactions.
