
Daniel Nelson
Professor
Nelson Group (240) 314-6249 nelsond@umd.eduRESEARCH INTERESTS
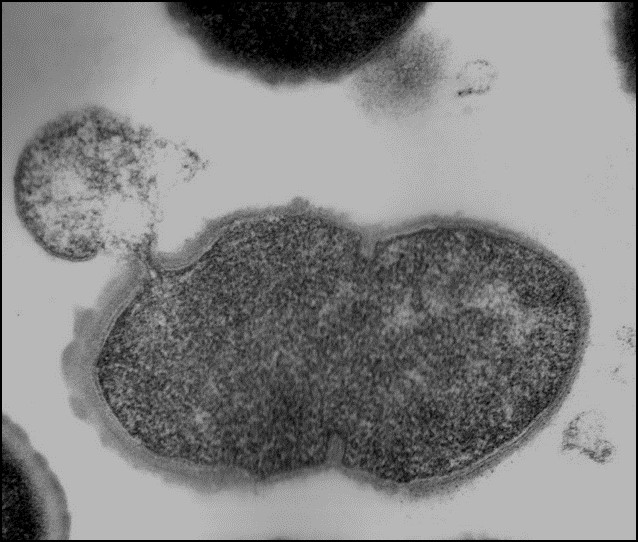
Dr. Nelson is an internationally recognized researcher in the field of antimicrobial discovery. The alarming increase of multidrug-resistant bacteria, the emergence of new pathogens, and the desire to reduce/eliminate antimicrobial use in agriculture products have prompted new antimicrobial discovery initiatives. Researchers seek to identify and develop alternative antimicrobial therapeutics that are not susceptible to traditional antibiotic resistance mechanisms. The Nelson lab harnesses peptidoglycan hydrolase enzymes, called endolysins, from bacteriophage and applies them to bacterial pathogens. These enzymes act rapidly on contact to degrade the bacterial cell wall of both animal and human pathogens, resulting in death of the bacterial cell.
CURRENT RESEARCH
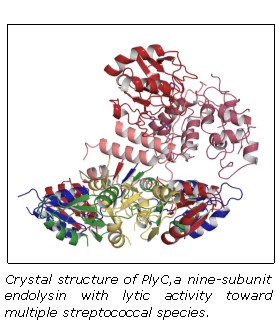
The Nelson lab works with endolysins effective against several human pathogens, including methicillin-resistant Staphylococcus aureus (i.e. MRSA), Clostridium difficile, Streptococcus pyogenes, and Bacillus anthracis (anthrax). The lab is also studying endolysins effective against critical animal pathogens, including Streptococcus equi (equine strangles disease), Streptococcus suis (meningitis and other infections in pigs), Streptococcus uberis (bovine mastitis), and Staphylococcus aureus (bovine mastitis).
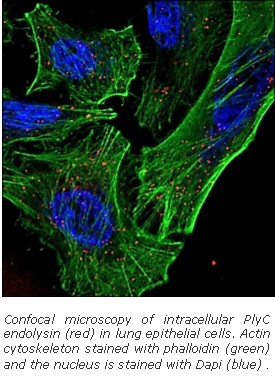
Dr. Nelson brings together the unique ability to cross disciplines and effectively incorporate cell biology, microbiology, and structural biology with bioengineering approaches to advance endolysin research and discovery. Employing rational methods (computational design or chimeragenesis) and random methods (directed evolution), his group is generating endolysins with more desirable attributes, such as higher activity, an expanded host range, a more favorable thermostability profile, or the ability to enter human cells to kill intracellular pathogens. It is anticipated that these bioengineering approaches will result in development of the next generation endolysins with enhanced properties.

Most recently, the Nelson lab is developing a platform technology, termed InstaVax, which exploits the ability of endolysin binding domains to target an “ImmunoBridge”, an antigen against which most people have circulating antibodies, to the surface of bacteria. The endolysin domains will redirect the pre-existing immunity against the ImmunoBridge towards the invading pathogen, leading to clearance of infection.
Publications
- Bacteriophage Endolysin treatment for systemic infection of Streptococcus iniae in hybrid striped bass.
- Thermal Characterization and Interaction of the Subunits from the Multimeric Bacteriophage Endolysin PlyC.
- A unique borrelial protein facilitates microbial immune evasion.
- Immunogenic epitope scanning in bacteriolytic enzymes Pal and Cpl-1 and engineering Pal to escape antibody responses.
- Understanding the Molecular Basis for Homodimer Formation of the Pneumococcal Endolysin Cpl-1.
- Bacteriophage endolysin powders for inhaled delivery against pulmonary infections.
- DNA Dye Sytox Green in Detection of Bacteriolytic Activity: High Speed, Precision and Sensitivity Demonstrated With Endolysins.
- Computational models in the service of X-ray and cryo-electron microscopy structure determination.
- Molecular basis for recognition of the Group A Carbohydrate backbone by the PlyC streptococcal bacteriophage endolysin.
- High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
- Application of bacteriophage-derived endolysins to combat streptococcal disease: current state and perspectives.
- Linker Editing of Pneumococcal Lysin ClyJ Conveys Improved Bactericidal Activity.
- Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus.
- Contributions of Net Charge on the PlyC Endolysin CHAP Domain.
- Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
- ClyJ Is a Novel Pneumococcal Chimeric Lysin with a Cysteine- and Histidine-Dependent Amidohydrolase/Peptidase Catalytic Domain.
- Safety Studies of Pneumococcal Endolysins Cpl-1 and Pal.
- Discovery and Biochemical Characterization of PlyP56, PlyN74, and PlyTB40-Bacillus Specific Endolysins.
- Borrelia burgdorferi surface-located Lmp1 protein processed into region-specific polypeptides that are critical for microbial persistence.
- Short communication: Recombinant bacteriophage endolysin PlyC is nontoxic and does not alter blood neutrophil oxidative response in lactating dairy cows.
- A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method.
- The Mga Regulon but Not Deoxyribonuclease Sda1 of Invasive M1T1 Group A Streptococcus Contributes to In Vivo Selection of CovRS Mutations and Resistance to Innate Immune Killing Mechanisms.
- Middle region of the Borrelia burgdorferi surface-located protein 1 (Lmp1) interacts with host chondroitin-6-sulfate and independently facilitates infection.
- Increasing the stability of the bacteriophage endolysin PlyC using rationale-based FoldX computational modeling.
- Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection.
- Quantitative analysis of the thermal stability of the gamma phage endolysin PlyG: a biophysical and kinetic approach to assaying therapeutic potential.
- Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A.
- Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus.
- Complete Genome Sequence of Staphylococcus aureus Phage GRCS.
- Crystal structure of ORF210 from E. coli O157:H1 phage CBA120 (TSP1), a putative tailspike protein.
- Determining carbapenemase activity with 18O labeling and targeted mass spectrometry.
- Purification and Characterization of Biofilm-Associated EPS Exopolysaccharides from ESKAPE Organisms and Other Pathogens.
- Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin.
