
Illarion Turko
Research Chemist
Turko Group (240) 314-6257 iturko@umd.eduProteins have multiple clinical applications--as biomarkers, therapeutics, and components of various biomaterials. All of these applications rely on accurate protein identification and quantification. Dr. Illarion Turko’s research focuses on the development of mass spectrometry measurements and protocols to quantitatively assess the concentrations of targeted proteins and their isoforms in clinically relevant biological samples.
CURRENT RESEARCH
Proteomic Toolbox to Standardize Purification of Extracellular Vesicles
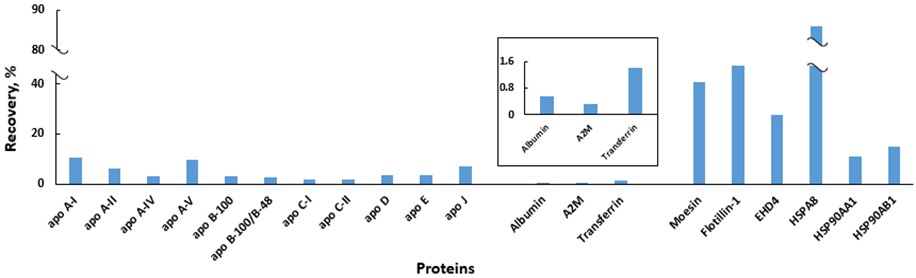
Extracellular vesicles (EVs) are stable membrane structures that can deliver their cargo remotely and regulate fundamental cellular responses, and EV-based therapeutics are heavily promoted by the biopharmaceutical industry. Pure EVs are needed to unambiguously define their functions; however, because of their low abundance, obtaining pure samples of EVs remains a challenge.
Dr. Turko’s group is working on the development of a quantitative mass spectrometry method to simultaneously measure concentrations of several groups of EV-specific proteins and non-EV proteins. They propose that this approach will provide a toolbox for evaluation of purification protocols to provide a better understanding of their prospects and limitations.
Assessing Morphology of Monoclonal Antibody (mAb) Aggregation
Many environmental factors can lead to aggregation of mAbs. The final state and form of aggregation seem to depend on the aggregation pathway. The extent to which different forms of mAb aggregates impact biological activity and the risk of immunogenicity is poorly understood, primarily because of the limitations of existing measurement techniques. Current techniques assess the size and number of aggregates, but not aggregate morphology. Dr. Turko’s team reasoned that protein-protein interfaces that are nonexistent in monomeric mAbs, but present in aggregated mAbs, could be targets for selective, high affinity, short peptide reagents.
This idea prompted the use of peptide phage display technology, a powerful tool in the identification of ligands with novel functions. Peptides bound to aggregate interfaces can be selected from a complex mixture of billions of displayed peptides on phage, and then further enriched through a ‘bio-panning’ process. Once identified, the selected peptides can be used for developing quantitative methods to assess the morphology of mAb aggregation. A proof-of-principle paper has been recently published (Cheung et al. 2017. Scientific Reports).
Publications
- QUANTITY: An Isobaric Tag for Quantitative Glycomics.
- Quantification of Borrelia burgdorferi Membrane Proteins in Human Serum: A New Concept for Detection of Bacterial Infection.
- Histone post-translational modifications in frontal cortex from human donors with Alzheimer's disease.
- Histone H3 Ser57 and Thr58 phosphorylation in the brain of 5XFAD mice.
- Quantification of histone deacetylase isoforms in human frontal cortex, human retina, and mouse brain.
- Quantitative performance of internal standard platforms for absolute protein quantification using multiple reaction monitoring-mass spectrometry.
- Quantifying CD4 receptor protein in two human CD4+ lymphocyte preparations for quantitative flow cytometry.
- Determining carbapenemase activity with 18O labeling and targeted mass spectrometry.
- Quantification of transferrin in human serum using both QconCAT and synthetic internal standards.
- Pretreatment with pyridoxamine mitigates isolevuglandin-associated retinal effects in mice exposed to bright light.
- Mass spectrometry quantification of PICALM and AP180 in human frontal cortex and neural retina.
- Mass spectrometry assessment of ubiquitin carboxyl-terminal hydrolase L1 partitioning between soluble and particulate brain homogenate fractions.
- Mass spectrometry quantification revealed accumulation of C-terminal fragment of apolipoprotein E in the Alzheimer's frontal cortex.
- Quantifying the cluster of differentiation 4 receptor density on human T lymphocytes using multiple reaction monitoring mass spectrometry.
- Quantification of amyloid precursor protein isoforms using quantification concatamer internal standard.
- 15N-labeled full-length apolipoprotein E4 as an internal standard for mass spectrometry quantification of apolipoprotein E isoforms.
- Mass spectrometry quantification of clusterin in the human brain.
- Expression and characterization of 15N-labeled human C-reactive protein in Escherichia coli and Pichia pastoris for use in isotope-dilution mass spectrometry.
- Sample prefractionation for mass spectrometry quantification of low-abundance membrane proteins.
- Features of the retinal environment which affect the activities and product profile of cholesterol-metabolizing cytochromes P450 CYP27A1 and CYP11A1.
- Quantification of cholesterol-metabolizing P450s CYP27A1 and CYP46A1 in neural tissues reveals a lack of enzyme-product correlations in human retina but not human brain.
- Optimizing the conditions of a multiple reaction monitoring assay for membrane proteins: quantification of cytochrome P450 11A1 and adrenodoxin reductase in bovine adrenal cortex and retina.
- Accumulation of large protein fragments in prematurely senescent ARPE-19 cells.
- Steroid and protein ligand binding to cytochrome P450 46A1 as assessed by hydrogen-deuterium exchange and mass spectrometry.
- Combined use of mass spectrometry and heterologous expression for identification of membrane-interacting peptides in cytochrome P450 46A1 and NADPH-cytochrome P450 oxidoreductase.
- Cyclic nucleotide phosphodiesterases: relating structure and function.
- Histidine-607 and histidine-643 provide important interactions for metal support of catalysis in phosphodiesterase-5.
- Phosphorylation of phosphodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic and allosteric cGMP-binding activities.
