
Roy Mariuzza
Professor
Mariuzza Group (240) 314-6243 rmariuzz@umd.eduResearch in Dr. Roy Mariuzza’s laboratory focuses on understanding how immune system cell surface receptors recognize molecules. Several classes of recognition molecules are under study: antibodies, T cell receptors (TCRs), natural killer (NK) cell receptors, and variable lymphocyte receptors (VLRs).
CURRENT RESEARCH
T cell recognition of self-antigens in multiple sclerosis
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system (CNS) driven by autoimmune CD4+ T cells. T cell responses to CNS self-antigens, such as myelin basic protein (MBP), are a critical autoimmune event in MS. To understand how T cells recognize self-antigens, the Mariuzza lab is determining structures of TCRs from MS patients bound to MBP peptides that are bound by MHC class II molecules.
T cell recognition of tumor antigens in human melanoma
Anti-tumor immunity can target non-mutated self-proteins or proteins with tumor-associated mutations or other modifications. The lab is carrying out structural studies using X-ray crystallography to investigate how tumor-specific CD4+ T cells from melanoma patients recognize each of these types of tumor antigens.
Structural basis for recognition of cellular and viral ligands by NK receptors
Natural killer (NK) cells are a first line of defense in immune responses against tumors and virally infected cells. A dynamic balance between activating receptors and inhibitory receptors regulates NK cell function. The lab is investigating the structural basis for NK receptor recognition of cellular and viral ligands using X-ray crystallography, and correlating this information with a better understanding of how NK cells function.
Evolution of the adaptive immune system
The evolutionary origin of adaptive immunity in vertebrates is the subject of much conjecture. The lab is studying the structural underpinnings of this process by defining the antigen recognition properties of newly discovered receptors expressed on the lymphocytes of jawless vertebrates (lamprey and hagfish). Antibodies are composed of Ig domains, but variable lymphocyte receptors (VLRs) consist of leucine-rich repeats. X-ray crystallographic studies of VLRs in complex with protein and carbohydrate antigens are providing unique insights into the diversity of molecular solutions nature has evolved for antigen recognition.
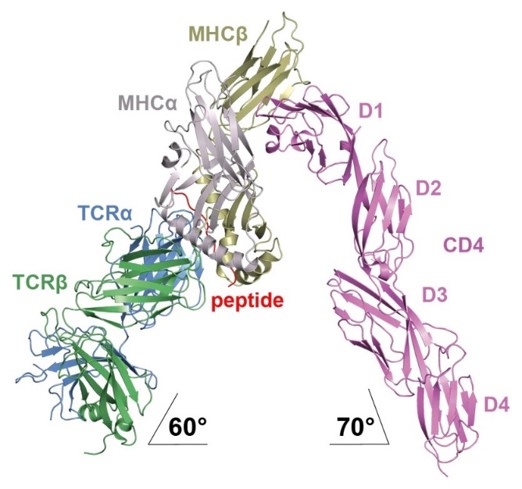
Structural analysis of the TCR–CD3 complex and TCR signaling
T cells are able to specifically recognize a peptide on the surface of another cell and then be activated to respond to this interaction. Peptides are short segments of protein molecules bound to a cell surface molecule called MHC. This peptide-MHC complex can engage a specific T cell receptor complex composed of a TCR heterodimer and CD3 molecules. The TCR mediates peptide–MHC (pMHC) recognition, while the CD3 molecules activate signals within the T cell. To understand the mechanism whereby TCR engagement by pMHC initiates signaling, the lab is investigating: 1) the spatial organization of the TCR–CD3 complex; and 2) potential structural changes in the TCR that are relayed to CD3. These studies are being undertaken through a combination of NMR spectroscopy, X-ray crystallography, and cell-based assays.
Structure-based design of hepatitis C vaccine
Hepatitis C virus (HCV) has devised multiple mechanisms of viral escape that contribute to the development of chronic infection. The lab is carrying out structural studies of binding interactions between human monoclonal antibodies and the HCV E2 envelope glycoprotein as well as structure-guided computational modeling to engineer E2 proteins that elicit broadly neutralizing anti-HCV antibodies for testing as vaccine candidates.
Publications
- Structural insights into clonal restriction and diversity in T cell recognition of two immunodominant SARS-CoV-2 nucleocapsid epitopes.
- Glycoengineering of the hepatitis C virus E2 glycoprotein improves biochemical properties and enhances immunogenicity.
- Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
- Glycoengineering of the hepatitis C virus E2 glycoprotein leads to improved biochemical properties and enhanced immunogenicity.
- Recognition of Self and Viral Ligands by NK Cell Receptors.
- Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
- Correction: Cutting Edge: LAG3 Dimerization Is Required for TCR/CD3 Interaction and Inhibition of Antitumor Immunity.
- Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
- Cutting Edge: LAG3 Dimerization Is Required for TCR/CD3 Interaction and Inhibition of Antitumor Immunity.
- The immune checkpoint receptor LAG3: Structure, function, and target for cancer immunotherapy.
- Structural basis for T cell recognition of cancer neoantigens and implications for predicting neoepitope immunogenicity.
- Zika virus NS4B protein targets TANK-binding kinase 1 and inhibits type I interferon production.
- CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
- Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
- Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
- Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum α-Glucosidase I with Anti-SARS-CoV-2 Activity.
- Cooperative binding of T cell receptor and CD4 to peptide-MHC enhances antigen sensitivity.
- Editorial overview: Engineering and design.
- Identification of Endoplasmic Reticulum α-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
- Induction of broadly neutralizing antibodies using a secreted form of the hepatitis C virus E1E2 heterodimer as a vaccine candidate.
- T cell receptors (TCRs) employ diverse strategies to target a p53 cancer neoantigen.
- Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
- N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum α-Glucosidases I and II with Antiviral Activity.
- Supramolecular assembly of Toll-like receptor 7/8 agonist into multimeric water-soluble constructs enables superior immune stimulation in vitro and in vivo.
- Design of a native-like secreted form of the hepatitis C virus E1E2 heterodimer.
- Crystal Structure of a Bivalent Antibody Fab Fragment.
- Peptide-MHC Binding Reveals Conserved Allosteric Sites in MHC Class I- and Class II-Restricted T Cell Receptors (TCRs).
- Structure-Based Design of Hepatitis C Virus E2 Glycoprotein Improves Serum Binding and Cross-Neutralization.
- In Vivo and In Vitro Potency of Polyphosphazene Immunoadjuvants with Hepatitis C Virus Antigen and the Role of Their Supramolecular Assembly.
- Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
- Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites.
- Identification of the fungal ligand triggering cytotoxic PRR-mediated NK cell killing of Cryptococcus and Candida.
- Insights into the Structural Basis of Antibody Affinity Maturation from Next-Generation Sequencing.
- Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope.
- Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8+ TCR Repertoires to Immunodominant Viral Antigens.
- The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis.
- Kinetic and thermodynamic studies of the interaction between activating and inhibitory Ly49 natural killer receptors and MHC class I molecules.
- Structural Insights into the Inhibitory Mechanism of an Antibody against B7-H6, a Stress-Induced Cellular Ligand for the Natural Killer Cell Receptor NKp30.
- Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design.
- Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice.
- Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain.
- Structural Basis for Clonal Diversity of the Public T Cell Response to a Dominant Human Cytomegalovirus Epitope.
- A positive cooperativity binding model between Ly49 natural killer cell receptors and the viral immunoevasin m157. KINETIC AND THERMODYNAMIC STUDIES.
- Pre-T-cell receptor binds MHC: Implications for thymocyte signaling and selection.
- Identification of the Docking Site for CD3 on the T Cell Receptor β Chain by Solution NMR.
- Expression, crystallization and X-ray diffraction analysis of a complex between B7-H6, a tumor cell ligand for the natural cytotoxicity receptor NKp30, and an inhibitory antibody.
- Peptidoglycan recognition protein-peptidoglycan complexes increase monocyte/macrophage activation and enhance the inflammatory response.
- Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody.
- Selection of the lamprey VLRC antigen receptor repertoire.
- Structural basis for recognition of cellular and viral ligands by NK cell receptors.
- T cell receptor bias for MHC: co-evolution or co-receptors?
- A positive cooperativity binding model between Ly49 natural killer cell receptors and the viral immunoevasin m157: kinetic and thermodynamic studies.
- Structure-based design of altered MHC class II-restricted peptide ligands with heterogeneous immunogenicity.
- Structural and biophysical insights into the role of CD4 and CD8 in T cell activation.
- Genetic Association of Peptidoglycan Recognition Protein Variants with Inflammatory Bowel Disease.
- Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
- Uptake and intracellular trafficking of superantigens in dendritic cells.
- Recognition of the Thomsen-Friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
- Structural insights into the evolution of the adaptive immune system.
- Structural basis for self-recognition by autoimmune T-cell receptors.
- Sugar-binding proteins from fish: selection of high affinity "lambodies" that recognize biomedically relevant glycans.
- Structural insights into the editing of germ-line-encoded interactions between T-cell receptor and MHC class II by Vα CDR3.
- Identification of multiple RIG-I-specific pathogen associated molecular patterns within the West Nile virus genome and antigenome.
- Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4.
- Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
- Cis-trans interactions of cell surface receptors: biological roles and structural basis.
- Structure of the human activating natural cytotoxicity receptor NKp30 bound to its tumor cell ligand B7-H6.
- Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection.
- The interaction with H-2D(d) in cis is associated with a conformational change in the Ly49A NK cell receptor.
- Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
- Assessing energetic contributions to binding from a disordered region in a protein-protein interaction .
- A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey.
- Structural insights into the evolution of the adaptive immune system: the variable lymphocyte receptors of jawless vertebrates.
- Structural basis for the presentation of tumor-associated MHC class II-restricted phosphopeptides to CD4+ T cells.
- The multiple mechanisms of T cell receptor cross-reactivity.
- Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis.
- High-affinity lamprey VLRA and VLRB monoclonal antibodies.
- Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen.
- Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
- Structural features of the full-length adaptor protein GADS in solution determined using small-angle X-ray scattering.
- Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors.
- Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
- A novel loop domain in superantigens extends their T cell receptor recognition site.
- TCR recognition of peptide/MHC class II complexes and superantigens.
- Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins.
- Superantigen natural affinity maturation revealed by the crystal structure of staphylococcal enterotoxin G and its binding to T-cell receptor Vbeta8.2.
- Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor.
- Peptidoglycan recognition proteins of the innate immune system.
- Variable dimerization of the Ly49A natural killer cell receptor results in differential engagement of its MHC class I ligand.
- Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule.
- Structural basis for recognition of MHC and MHC-like ligands by natural killer cell receptors.
- Crystal structure of human peptidoglycan recognition protein I alpha bound to a muramyl pentapeptide from Gram-positive bacteria.
- Multiple paths to multispecificity.
- Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
- Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs).
- Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
- Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins I{alpha} and S.
- Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
- Structural basis for recognition of the T cell adaptor protein SLP-76 by the SH3 domain of phospholipase Cgamma1.
- Binding of natural variants of staphylococcal superantigens SEG and SEI to TCR and MHC class II molecule.
- Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s).
- Crystal structure of a peptidoglycan recognition protein (PGRP) in complex with a muramyl tripeptide from Gram-positive bacteria.
- Crystal structure of human peptidoglycan recognition protein S (PGRP-S) at 1.70 A resolution.
- Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces.
- Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation.
- Structural basis for peptidoglycan binding by peptidoglycan recognition proteins.
- Entropically assisted carbohydrate recognition by a natural killer cell-surface receptor.
- Cloning, expression and interaction of human T-cell receptors with the bacterial superantigen SSA.
- Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha.
- Structural basis for differential recognition of tyrosine-phosphorylated sites in the linker for activation of T cells (LAT) by the adaptor Gads.
- Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition.
- Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
- Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
- Dissecting cooperative and additive binding energetics in the affinity maturation pathway of a protein-protein interface.
- Expression, crystallization and preliminary crystallographic analysis of the extracellular IgV-like domain of the human natural killer cell inhibitory receptor p75/AIRM1.
- Structural, energetic, and functional analysis of a protein-protein interface at distinct stages of affinity maturation.
- Exploration of the P6/P7 region of the peptide-binding site of the human class II major histocompatability complex protein HLA-DR1.
- Functional analysis of the TCR binding domain of toxic shock syndrome toxin-1 predicts further diversity in MHC class II/superantigen/TCR ternary complexes.
- A growing family of natural killers.
- Combined affinity and rate constant distributions of ligand populations from experimental surface binding kinetics and equilibria.
- X-ray snapshots of the maturation of an antibody response to a protein antigen.
- Conservation of nonpeptide antigen recognition by rhesus monkey V gamma 2V delta 2 T cells.
- Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen.
- Molecular recognition in antibody-antigen complexes.
- Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
- Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line.
- Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
- MHC class I recognition by Ly49 natural killer cell receptors.
- Quantifying the energetics of cooperativity in a ternary protein complex.
- Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination.
- Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster.
- So many ways of getting in the way: diversity in the molecular architecture of superantigen-dependent T-cell signaling complexes.
- Binding of the natural killer cell inhibitory receptor Ly49A to its major histocompatibility complex class I ligand. Crucial contacts include both H-2Dd AND beta 2-microglobulin.
- A response calculus for immobilized T cell receptor ligands.
- Structural basis of MHC class I recognition by natural killer cell receptors.
- Structural features of nonpeptide prenyl pyrophosphates that determine their antigenicity for human gamma delta T cells.
- Role of the T cell receptor ligand affinity in T cell activation by bacterial superantigens.
- High affinity T cell receptors from yeast display libraries block T cell activation by superantigens.
- Superantigen recognition by gammadelta T cells: SEA recognition site for human Vgamma2 T cell receptors.
- MHC class I molecules, structure and function.
- Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
- Mapping the ligand of the NK inhibitory receptor Ly49A on living cells.
- Antigen recognition by human gamma delta T cells: pattern recognition by the adaptive immune system.
- Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
- Crystal structure of human CD69: a C-type lectin-like activation marker of hematopoietic cells.
- Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
- Luxury accommodations: the expanding role of structural plasticity in protein-protein interactions.
- Three-dimensional structures of the free and antigen-bound Fab from monoclonal antilysozyme antibody HyHEL-63(,).
- Mapping the energy of superantigen Staphylococcus enterotoxin C3 recognition of an alpha/beta T cell receptor using alanine scanning mutagenesis.
