Profile

Travis Gallagher
Research Chemist
Gallagher Group (240) 314-6204 dgallag1@umd.edu
Dr. Travis Gallagher’s research combines crystallography with other measurements and modeling strategies to provide structural descriptions of proteins and their interactions. Three major current foci are antibodies, vaccines and T cell receptors.
CURRENT RESEARCH
Antibody Structure
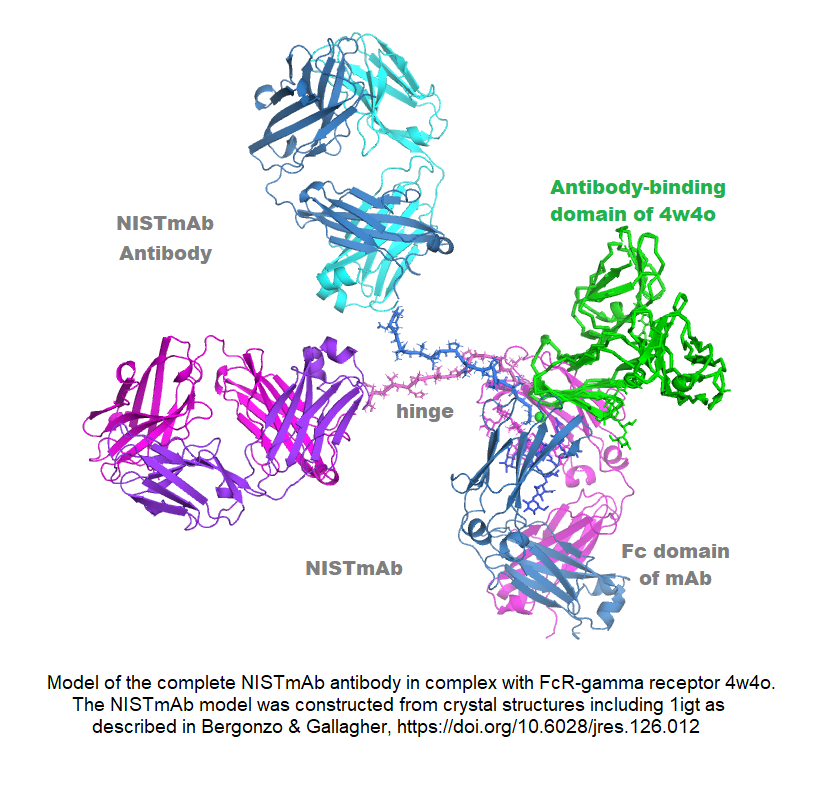
The NIST standard antibody (reference material 8671, also called NISTmAb), has become a widely used industry standard, largely because of its extensive characterization at IBBR and at NIST. Dr. Gallagher has produced crystal structures of both the Fab and Fc fragments, and combined them with other information to generate an all-atom structural model of this IgG1 antibody. The pdb format model contains all 1326 amino acids in four chains, plus complete G1F/G0F glycans, and is available from data.nist.gov, along with the associated publication. The model is useful for conformational study of its biological signaling complexes (see Figure) and as a starting point for molecular dynamics. A long-term goal is to map the conformational space of the antibody to better understand its range and dimensionality, and to address the relation between conformation and biologically important phenomena like aggregation and signaling.
Vaccines and T Cell Receptors
Bacterial proteins tend to stimulate the human immune system and have been used as components in several successful vaccine constructs. With material provided by a local vaccine research and development company, Dr. Gallagher has solved the first structure of a monomeric CRM-197 from Corynebacterium diphtheriae. The structure enables modeling of the dimerization process and will be useful for carrier-based vaccine strategies generally.
T cell receptors (TCR) that bind to antigens from SARS-CoV-2 are the focus of a current ‘Seed Grant’ collaboration among IBBR researchers Mariuzza, Pierce, and Gallagher. In this work, crystal structures of TCR complexes with viral epitopes presented by MHC molecules are determined in order to advance understanding of how T cells contribute to defenses against this pandemic virus. While dozens of antibodies that bind the SARS-CoV-2 spike protein have been structurally characterized, comparatively little is known about the corresponding T cell responses, which include reactions against non-spike proteins from within the virus.
Publications
- Effects of glycans and hinge on dynamics in the IgG1 Fc.
- SARS-CoV-2 infection establishes a stable and age-independent CD8+ T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
- Monomeric crystal structure of the vaccine carrier protein CRM197 and implications for vaccine development.
- Design and characterization of a protein fold switching network.
- Reversible switching between two common protein folds in a designed system using only temperature.
- Structure of the Fc fragment of the NIST reference antibody RM8671.
- Quantitative analysis of the impact of a human pathogenic mutation on the CCT5 chaperonin subunit using a proxy archaeal ortholog.
- Characterization of the NISTmAb Reference Material using small-angle scattering and molecular simulation : Part I: Dilute protein solutions.
