Profile

Alexander MacKerell
Dr. Alexander MacKerell’s research involves the development and application of computational methods to investigate the relationships of structure and dynamics to function in a range of biological and chemical systems. These efforts include empirical force field development, implementation of novel sampling methodologies, understanding the physical forces driving the structure and dynamics of proteins, nucleic acids, and carbohydrates, and computer-aided drug design (CADD) studies and methodology development. The MacKerell lab works closely with experimentalists in the area of drug development to provide detailed interpretation of experimental data while simultaneously refining and developing novel theoretical approaches.
The MacKerell lab is responsible for developing and maintaining the empirical force fields used in a number of simulation packages including the program CHARMM (Chemistry at HARvard Macromolecular Mechanics). CADD methodologies developed in the MacKerell lab include SILCS (Site-Identification by Ligand Competitive Saturation) and CSP (Conformationally Sampled Pharmacophore).
CURRENT RESEARCH
A central focus of Dr. MacKerell’s group is the continued development and extension of empirical force fields and enhanced sampling methods for use in the simulation of biological macromolecules. Such simulations are important in advancing the understanding of how intrinsic characteristics and environmental conditions contribute to the conformational properties of proteins, nucleic acids, lipids, carbohydrates, and small molecules.
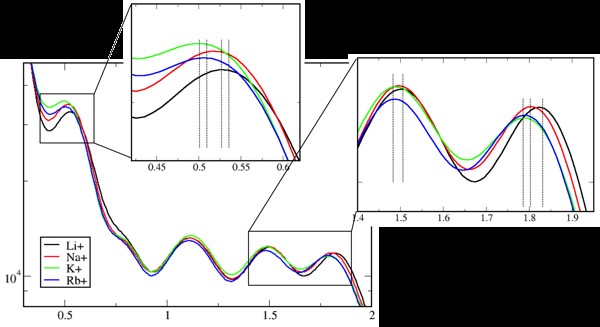
In addition to widely used additive force fields, the MacKerell lab developed a polarizable force field for biomolecules, based on a classical Drude oscillator model. Drude-2013 extends the capabilities of molecular dynamics simulations, as in the example shown in the graph to the right.
CADD is a powerful tool that must contend with both the large conformational space of flexible macromolecular drug targets like proteins and the large chemical space of potential drug-like molecules to be screened against the target. Work in the MacKerell lab seeks to address these challenges with the development of new methodologies. For example, the SILCS (Site Identification by Ligand Competitive Saturation) methodology may be used for lead compound identification and optimization, evaluation of protein-protein interactions, and optimization of formulation via rational selection of excipients and buffer.
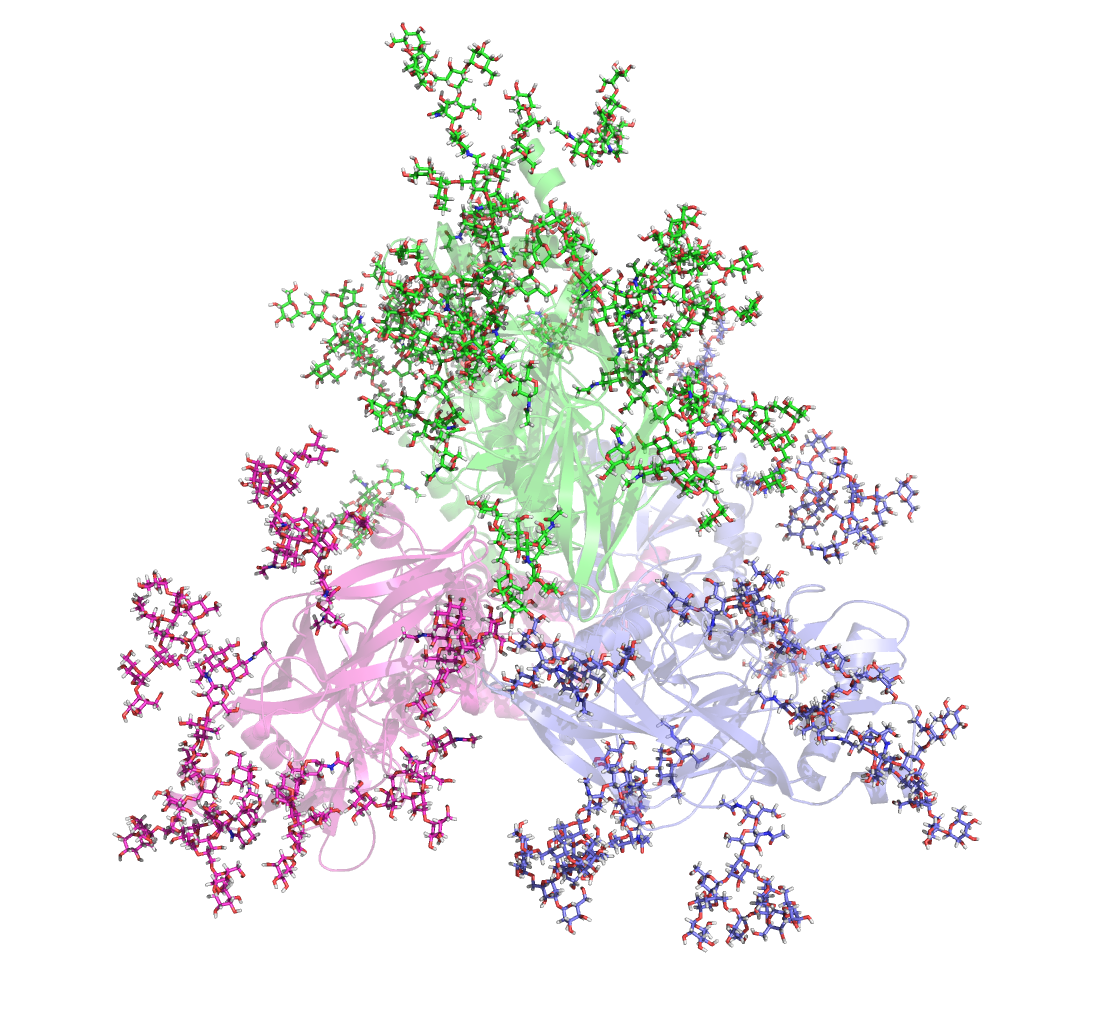
Additional activities in the MacKerell lab include structure-function studies of carbohydrates, proteins, and nucleic acids. Recently, the lab used enhanced sampling methods to explore the conformational heterogeneity HIV envelope protein glycans (Yang et al. 2017, Scientific Reports), resulting in the identification of novel glycan-antibody interactions that may play a role in neutralizing the virus. See https://mackerell.umaryland.edu/ for additional information on the MacKerell lab.
Publications
- Computational Modeling of PROTAC Ternary Complexes as Ensembles Using SILCS-xTAC.
- Arrhythmia Risk Predictions from Molecular Simulations of Cardiac Ion Channel-Drug Interactions.
- Drude Polarizable Force Field for Phosphorylated Polypeptides and Proteins.
- Prediction of TdP Arrhythmia Risk Through Molecular Simulations of Conformation-specific Drug Interactions with the hERG K + , Na V 1.5, and Ca V 1.2 Channels.
- Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors.
- The mammalian SKI complex is a broad-spectrum antiviral drug target that upregulates cellular cholesterol to inhibit viral replication.
- GaSal-2: A Water-Soluble Antipseudomonal Agent Targeting the Extracellular Hemophore HasAp.
- Harnessing computational technologies to facilitate antibody-drug conjugate development.
- Non-Covalent Molecular Interaction Rules to Define Internal Dimer Coordinates for Quantum Mechanical Potential Energy Scans.
- High-Throughput Ligand Dissociation Kinetics Predictions Using Site Identification by Ligand Competitive Saturation.
- The need to implement FAIR principles in biomolecular simulations.
- Potent and Selective Human Constitutive Androstane Receptor Activator DL5055 Facilitates Cyclophosphamide-Based Chemotherapies.
- Mapping the Distribution and Affinities of Ligand Interaction Sites on Human Serum Albumin.
- Increasing the Accuracy and Robustness of the CHARMM General Force Field with an Expanded Training Set.
- Investigating the Interaction between Excipients and Monoclonal Antibodies PGT121 and N49P9.6-FR-LS: A Comprehensive Analysis.
- First-in-class mitogen-activated protein kinase (MAPK) p38α: MAPK-activated protein kinase 2 dual signal modulator with anti-inflammatory and endothelial-stabilizing properties.
- Computationally Efficient Polarizable MD Simulations: A Simple Water Model for the Classical Drude Oscillator Polarizable Force Field.
- Detection of Putative Ligand Dissociation Pathways in Proteins Using Site-Identification by Ligand Competitive Saturation.
- Grand canonical Monte Carlo and deep learning assisted enhanced sampling to characterize the distribution of Mg2+ and influence of the Drude polarizable force field on the stability of folded states of the twister ribozyme.
- Modeling Ligand Binding Site Water Networks with Site Identification by Ligand Competitive Saturation: Impact on Ligand Binding Orientations and Relative Binding Affinities.
- Isolation of phytoconstituents from an extract of Murraya paniculata with cytotoxicity and antioxidant activities and in silico evaluation of their potential to bind to aldose reductase (AKR1B1).
- Balancing Group 1 Monoatomic Ion-Polar Compound Interactions in the Polarizable Drude Force Field: Application in Protein and Nucleic Acid Systems.
- Revised 4-Point Water Model for the Classical Drude Oscillator Polarizable Force Field: SWM4-HLJ.
- Refinement of the Drude Polarizable Force Field for Hexose Monosaccharides: Capturing Ring Conformational Dynamics with Enhanced Accuracy.
- Exploring Druggable Binding Sites on the Class A GPCRs Using the Residue Interaction Network and Site Identification by Ligand Competitive Saturation.
- CHARMM at 45: Enhancements in Accessibility, Functionality, and Speed.
- First-in-Class Mitogen-Activated Protein Kinase p38α: MAPK-Activated Protein Kinase-2 (MK2) Dual Signal Modulator with Anti-inflammatory and Endothelial-stabilizing Properties.
- Combined Physics- and Machine-Learning-Based Method to Identify Druggable Binding Sites Using SILCS-Hotspots.
- Enhancing SILCS-MC via GPU Acceleration and Ligand Conformational Optimization with Genetic and Parallel Tempering Algorithms.
- FFParam-v2.0: A Comprehensive Tool for CHARMM Additive and Drude Polarizable Force-Field Parameter Optimization and Validation.
- Identifying and Assessing Putative Allosteric Sites and Modulators for CXCR4 Predicted through Network Modeling and Site Identification by Ligand Competitive Saturation.
- Balancing Group I Monatomic Ion-Polar Compound Interactions for Condensed Phase Simulation in the Polarizable Drude Force Field.
- Biomolecular dynamics in the 21st century.
- Dendritic Cell-Mediated Cross-Priming by a Bispecific Neutralizing Antibody Boosts Cytotoxic T Cell Responses and Protects Mice against SARS-CoV-2.
- Non-β Lactam Inhibitors of the Serine β-Lactamase blaCTX-M15 in Drug-Resistant Salmonella typhi.
- Combining SILCS and Artificial Intelligence for High-Throughput Prediction of the Passive Permeability of Drug Molecules.
- Nitro-benzylideneoxymorphone, a bifunctional mu and delta opioid receptor ligand with high mu opioid receptor efficacy.
- Identification of a novel transport system in Borrelia burgdorferi that links the inner and outer membranes.
- Influence of Mg2+ Distribution on the Stability of Folded States of the Twister Ribozyme Revealed Using Grand Canonical Monte Carlo and Generative Deep Learning Enhanced Sampling.
- Integrated Covalent Drug Design Workflow Using Site Identification by Ligand Competitive Saturation.
- GPU-specific algorithms for improved solute sampling in grand canonical Monte Carlo simulations.
- Drude Polarizable Lipid Force Field with Explicit Treatment of Long-Range Dispersion: Parametrization and Validation for Saturated and Monounsaturated Zwitterionic Lipids.
- Site Identification by Ligand Competitive Saturation-Biologics Approach for Structure-Based Protein Charge Prediction.
- Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum α-Glucosidase I with Anti-SARS-CoV-2 Activity.
- hERG Blockade Prediction by Combining Site Identification by Ligand Competitive Saturation and Physicochemical Properties.
- Glycosidic α-linked mannopyranose disaccharides: an NMR spectroscopy and molecular dynamics simulation study employing additive and Drude polarizable force fields.
- In silico identification of a β2-adrenoceptor allosteric site that selectively augments canonical β2AR-Gs signaling and function.
- Computer-Aided Drug Design: An Update.
- Scaffold hopping from indoles to indazoles yields dual MCL-1/BCL-2 inhibitors from MCL-1 selective leads.
- Extension of the CHARMM Classical Drude Polarizable Force Field to N- and O-Linked Glycopeptides and Glycoproteins.
- Spatial requirements for ITAM signaling in an intracellular natural killer cell model membrane.
- Preserving the Integrity of Empirical Force Fields.
- SILCS-RNA: Toward a Structure-Based Drug Design Approach for Targeting RNAs with Small Molecules.
- Accurate modeling of RNA hairpins through the explicit treatment of electronic polarizability with the classical Drude oscillator force field.
- In Silico Identification of a β2 Adrenergic Receptor Allosteric Site that Selectively Augments Canonical β2 AR-Gs Signaling and Function.
- Cholecalciferol complexation with hydroxypropyl-β-cyclodextrin (HPBCD) and its molecular dynamics simulation.
- Harnessing Deep Learning for Optimization of Lennard-Jones Parameters for the Polarizable Classical Drude Oscillator Force Field.
- Application of Site-Identification by Ligand Competitive Saturation in Computer-Aided Drug Design.
- Deep Neural Network Model to Predict the Electrostatic Parameters in the Polarizable Classical Drude Oscillator Force Field.
- Computational and Experimental Characterization of rDNA and rRNA G-Quadruplexes.
