Profile

Christina Bergonzo
Dr. Bergonzo develops and applies molecular dynamics simulations to study biomolecular structure prediction. Current work focuses on extending molecular dynamics force fields to address chemical modifications to biotherapeutic RNA, and combining simulation with new experimental data, including small and wide angle X-ray scattering and NMR.
https://www.nist.gov/people/christina-bergonzo
Publications
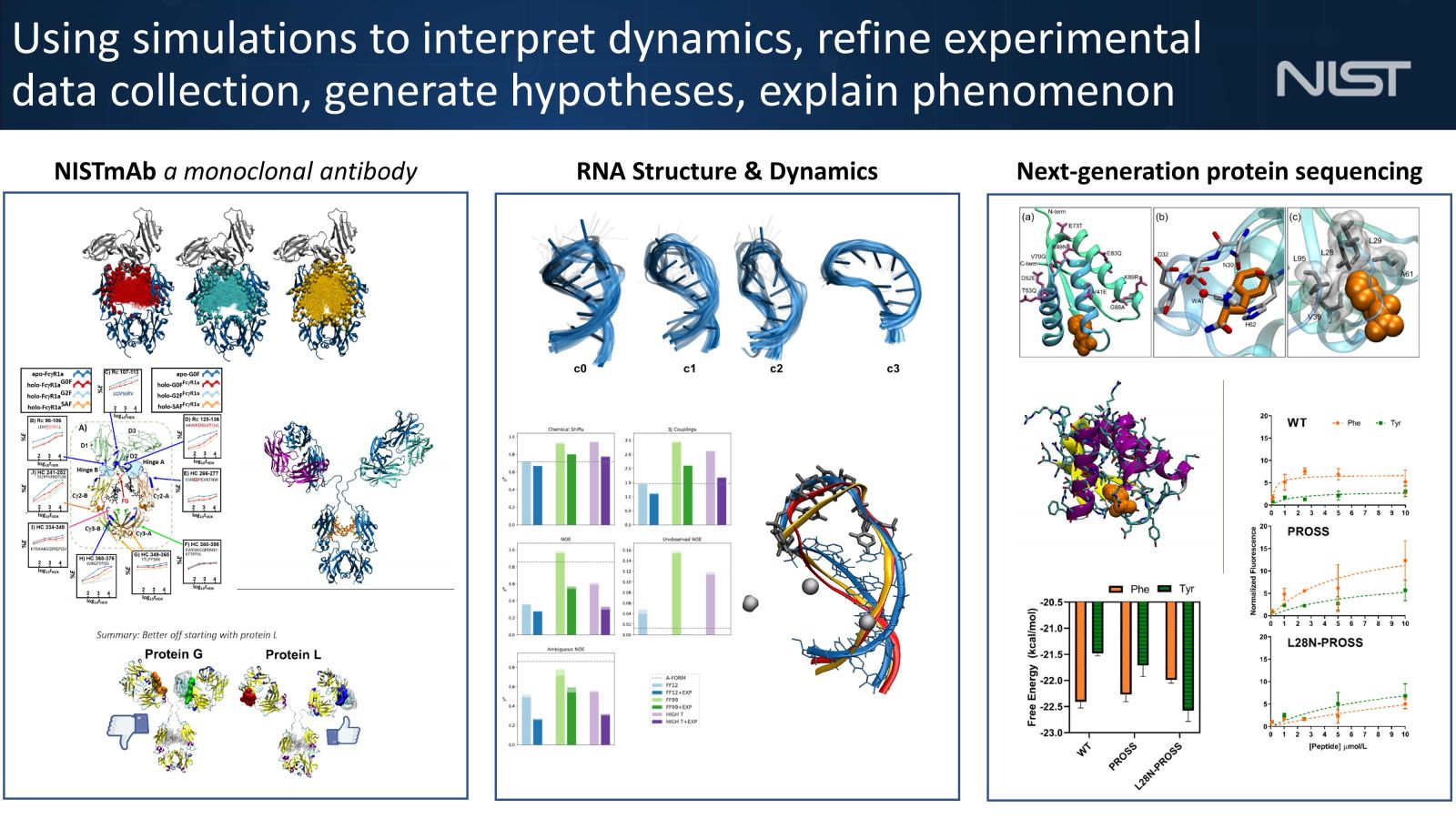
- Spatial and Sequential Topological Analysis of Molecular Dynamics Simulations of IgG1 Fc Domains.
- Critical Assessment of RNA and DNA Structure Predictions via Artificial Intelligence: The Imitation Game.
- Structure and dynamics of monoclonal antibody domains determined using spins, scattering, and simulations.
- Conformational dynamics of the hepatitis B virus pre-genomic RNA on multiple time scales: implications for viral replication.
- Conformational Heterogeneity of UCAAUC RNA Oligonucleotide from Molecular Dynamics Simulations, SAXS, and NMR experiments.
- prepareforleap: An automated tool for fast PDB-to-parameter generation.
- Characterization of the internal translation initiation region in monoclonal antibodies expressed in Escherichia coli.
- Accuracy of MD solvent models in RNA structure refinement assessed via liquid-crystal NMR and spin relaxation data.
- Structural insights into DNA-stabilized silver clusters.
- Maximizing accuracy of RNA structure in refinement against residual dipolar couplings.
- Mg2+ Binding Promotes SLV as a Scaffold in Varkud Satellite Ribozyme SLI-SLV Kissing Loop Junction.
- Investigating the ion dependence of the first unfolding step of GTPase-Associating Center ribosomal RNA.
- DNA Deformation-Coupled Recognition of 8-Oxoguanine: Conformational Kinetic Gating in Human DNA Glycosylase.
- Improved Force Field Parameters Lead to a Better Description of RNA Structure.
- A dynamic checkpoint in oxidative lesion discrimination by formamidopyrimidine-DNA glycosylase.
- Stem-Loop V of Varkud Satellite RNA Exhibits Characteristics of the Mg(2+) Bound Structure in the Presence of Monovalent Ions.
- Highly sampled tetranucleotide and tetraloop motifs enable evaluation of common RNA force fields.
- Molecular modeling of nucleic Acid structure: electrostatics and solvation.
- Molecular modeling of nucleic Acid structure: setup and analysis.
- Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition.
- Evaluation of enhanced sampling provided by accelerated molecular dynamics with Hamiltonian replica exchange methods.
- Molecular modeling of nucleic acid structure.
- Molecular modeling of nucleic acid structure: energy and sampling.
- Multidimensional Replica Exchange Molecular Dynamics Yields a Converged Ensemble of an RNA Tetranucleotide.
