Profile

Edwin Pozharski
(he/him)
(240) 314-6870 epozhars@umd.eduDr. Pozharskiy's area of scientific expertise is structural biology, with a major focus on protein X-ray crystallography. Throughout his career as a protein crystallographer, Dr. Pozharskiy has focused on structural mechanisms of molecular recognition in a range of biomolecular systems, including small molecules, protein-DNA, and protein-protein interactions. He has also contributed to methodological aspects of protein crystallography, including computational methods, structure validation, and crystallization methods. As a molecular biophysicist, Dr. Pozharskiy has expertise in membrane biophysics and protein thermodynamics, including the study of cationic lipids and their DNA complexes. He utilizes a variety of biophysical techniques, such as dynamic light scattering, protein fluorescence and fluorescence anisotropy, differential scanning fluorimetry, and isothermal titration calorimetry.
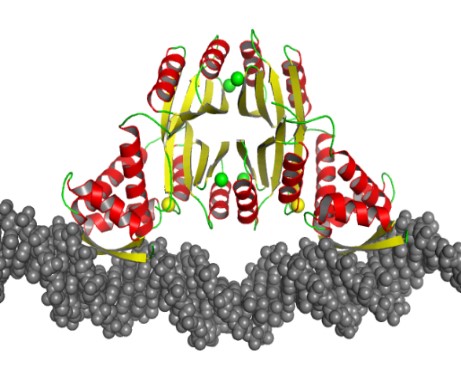
As Structural Biology co-Section Leader of the Center for Biomolecular Therapeutics (CBT), Dr. Pozharskiy is involved in various projects focused on structure-based drug design against therapeutic targets related to cancer, diabetes, and infectious disease. He is also involved in a recent joint initiative by IBBR partner institutions to establish research capabilities in single-particle cryoelectron microscopy.
CURRENT RESEARCH
Structural studies of DNA recognition by transcription factors and enzymes
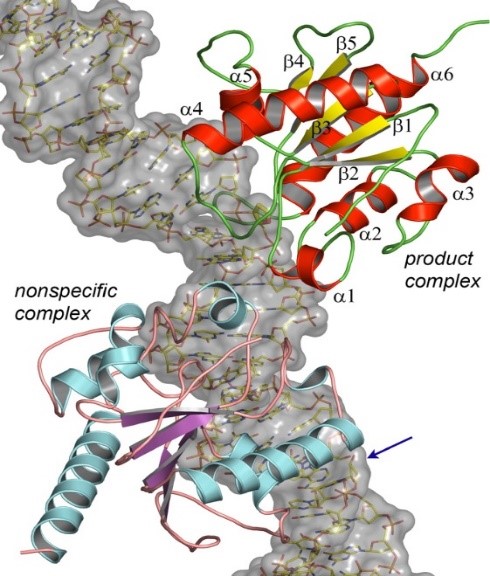
Dr. Pozharskiy’s work on the molecular recognition of DNA by protein molecules includes structural studies of the nickel-dependent transcription factor NikR from Helicobacter pylori. This protein is essential for the bacterium’s ability to survive acidic environments during colonization, as it activates a secreted urease enzyme that is capable of lowering local pH. Previous studies in collaboration with Sara Michel, Professor, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, have revealed the unique mode of metal binding by this protein. A recently solved crystal structure of the NikR/DNA complex explains the evolutionary mechanism of this transcription factor’s broad specificity to multiple DNA targets.
Another area of research includes structural studies of DNA binding and the evolutionary underpinnings of substrate recognition by human thymine specific DNA glycosylase, an essential enzyme in DNA repair and modification arising in epigenetic context. This work is a collaboration with Alexander Drohat, Professor, Department of Biochemistry & Molecular Biology, University of Maryland School of Medicine.
Structural studies of binary toxin from Clostridium difficile
Clostridium difficile is an opportunistic human pathogen. A recently emerging, highly virulent strain of this bacterium is characterized by the expression of binary toxin CDT, in addition to two canonical C. difficile toxins, TcdA and TcdB. A team-based project in the CBT currently aims to develop novel treatments against this toxin. In collaboration with Amédée des Georges, Assistant Professor of Chemistry & Biochemistry, City College of New York, efforts are currently underway to structurally characterize this binary toxin using single-particle cryoelectron microscopy, focusing on the large oligomeric cell binding component. The enzymatic component of the binary toxin is being targeted for drug design using protein crystallography.
Publications
- Rational design of flavivirus E protein vaccine optimizes immunogenicity and mitigates antibody dependent enhancement risk.
- Pore formation by the CDTb component of the Clostridioides difficile binary toxin is Ca2+-dependent.
- A comprehensive engineering strategy improves potency and manufacturability of a near pan-neutralizing antibody against HIV.
- A comprehensive engineering strategy improves potency and manufacturability of a near pan-neutralizing antibody against HIV.
- Structural and Functional Insights into the Delivery Systems of Bacillus and Clostridial Binary Toxins.
- Directly visualizing individual polyorganophosphazenes and their single-chain complexes with proteins.
- Cryo-EM and AFM visualize linear polyorganophosphazene: individual chains and single-chain assemblies with proteins.
- Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
- Dendritic Cell-Mediated Cross-Priming by a Bispecific Neutralizing Antibody Boosts Cytotoxic T Cell Responses and Protects Mice against SARS-CoV-2.
- Endosomal activation of the Clostridioides difficile binary toxin is Ca 2+ -dependent.
- Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
- Structure-function Analyses Reveal Key Molecular Determinants of HIV-1 CRF01_AE Resistance to the Entry Inhibitor Temsavir.
- Characterizing inhibitors of human AP endonuclease 1.
- Detect, correct, retract: How to manage incorrect structural models.
- Validation of Protein-Ligand Crystal Structure Models: Small Molecule and Peptide Ligands.
- Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
- X-ray crystal structure of human calcium-bound S100A1.
- Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif.
- Twilight reloaded: the peptide experience.
- Short Carboxylic Acid-Carboxylate Hydrogen Bonds Can Have Fully Localized Protons.
- Fluorescence Resonance Energy Transfer (FRET)-Based Analysis of Lipoplexes.
- Fluorometric Analysis of Individual Cationic Lipid-DNA Complexes.
- Structural Basis for Excision of 5-Formylcytosine by Thymine DNA Glycosylase.
- Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
- A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
- Additional Comment on Three X-ray Crystal Structure Papers.
- Comment on Three X-ray Crystal Structure Papers.
- Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg2+ cofactor.
- Structural and functional analysis of the pro-domain of human cathelicidin, LL-37.
- Visualizing ligand molecules in Twilight electron density.
- Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures.
- Structure of extracellular signal-regulated kinase 2 in complex with ATP and ADP.
- Novel structural features of xylanase A1 from Paenibacillus sp. JDR-2.
- On the variability of experimental data in macromolecular crystallography.
- How a mismatch repair enzyme balances the needs for efficient lesion processing and minimal action on undamaged DNA.
- Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA.
- Ni(II) coordination to mixed sites modulates DNA binding of HpNikR via a long-range effect.
- Rapid Crystallization of L-Alanine on Engineered Surfaces using Metal-Assisted and Microwave-Accelerated Evaporative Crystallization.
- Apparent instability of crystallographic refinement in the presence of disordered model fragments and upon insufficiently restrained model geometry.
- N-Acetylfarnesylcysteine is a novel class of peroxisome proliferator-activated receptor γ ligand with partial and full agonist activity in vitro and in vivo.
- Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.
- In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
- Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
- A conserved protein interaction interface on the type 5 G protein beta subunit controls proteolytic stability and activity of R7 family regulator of G protein signaling proteins.
- Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups.
- Holo-Ni(II)HpNikR is an asymmetric tetramer containing two different nickel-binding sites.
- Percentile-based spread: a more accurate way to compare crystallographic models.
- Studies on ligand binding to histidine triad nucleotide binding protein 1.
- The effects of CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography.
- Fluorescence resonance energy transfer-based analysis of lipoplexes.
- Fluorometric Analysis of Individual Cationic Lipid-DNA Complexes.
- Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
- Crystallization and crystallographic analysis of Bacillus subtilis xylanase C.
- The role of bias in crystallization conditions in automated microseeding.
- Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
- Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
- Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
