Profile

Ella Mihailescu
Associate Research Professor
Mihailescu Group (240) 314-6123 ella11@umd.eduDr. Ella Mihailescu’s research is focused on developing biophysical methods for investigations of the structural interactions of membrane proteins, membrane-active peptides, and lipophilic drug molecules with lipid membranes. A major effort in the Mihailescu laboratory is directed toward advancing precision measurement of membrane protein structures in engineered lipid platforms.
CURRENT RESEARCH
The Mihailescu lab utilizes their unique experimental expertise in neutron and X-ray scattering methods, surface-sensitive techniques, and computational approaches to solve challenging problems in the area of membrane protein (MP) research. MPs are critically important to the pharmaceutical industry, as they are the target of half of all drugs; 33% target G-protein-coupled receptors and 18% target membrane ion channels (Santos et al. 2017. Nat. Rev. Drug. Discov.). Macroscopic observations from cell-based assays and empirical approaches in drug discovery must be informed by molecular level information obtained with structural techniques.
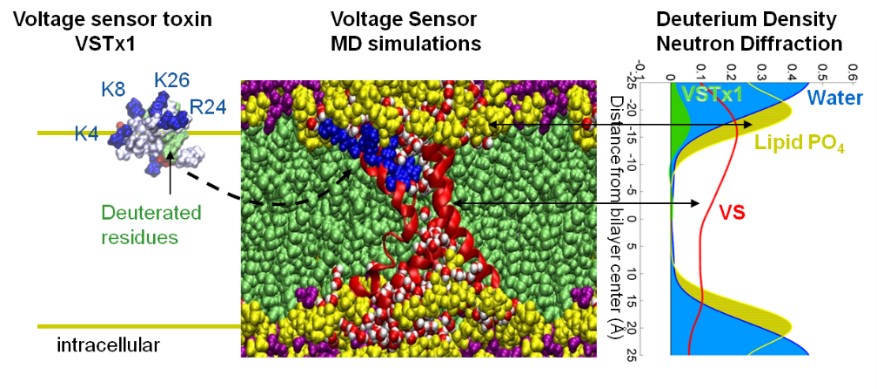
Neutron Diffraction Techniques for Measurements of Lipid Membranes and Membrane Protein Structures
Interactions between a membrane protein and its surrounding lipid bilayer can be uniquely inferred from neutron diffraction patterns of deuterium-labeled samples. To this end, the lab is exploring methods for functional reconstitution of membrane proteins and peptides in conjunction with deuterium labeling. The diffraction measurements can be complemented computationally with molecular dynamics (MD) simulations. For instance, diffraction data led to information about the conformations, and interactions that ion channel proteins and gating-modifier toxins from tarantula venom (e.g. VSTx1) establish in the bilayer environment. This research helped to resolve controversial issues regarding mechanistic models of voltage-gated ion channel activation.
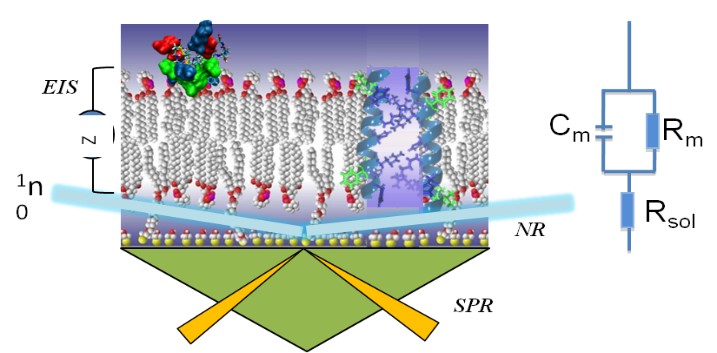
Surface Sensitive Techniques
Membrane-active peptides and lipophilic drugs (e.g. antimicrobial peptides, toxins, hormones, NSAIDs) work by binding to and entering bilayer environments, causing membrane deformations and changes in the electrical properties of the bilayer. For many pore-forming toxins and antimicrobial peptides, interactions with the bilayer lipids is a key event that drives folding into functional secondary structures and assembly of peptide monomers into transmembrane pores. Electrical Impedance Spectroscopy (EIS) can exquisitely capture the onset of membrane permeabilization through such protein pores or channels. Dr. Mihailescu seeks to understand what properties of antimicrobial peptides make them potent antibiotics by coupling structural and functional observations of their behavior in membranes. Her goal is to discover novel antimicrobial therapeutics to combat drug-resistant bacteria. This work is a joint effort with Dr. Vitalii Silin, IBBR Fellow and Associate Research Professor, and Dr. Frank Heinrich, NIST Center for Neutron Research.
