Profile

Jaekyun Jeon
Assistant Research Professor
Jeon Group (240) 314-6389 jkjeon@umd.eduDr. Jaekyun Jeon’s laboratory focuses on the development and the application of biophysical methods to characterize the structures and molecular mechanisms of biomolecules. The primary method is solid-state NMR, which provides structural details at atomic level for a wide range of samples including biomolecules at insoluble, lyophilized, or frozen states.
Current research
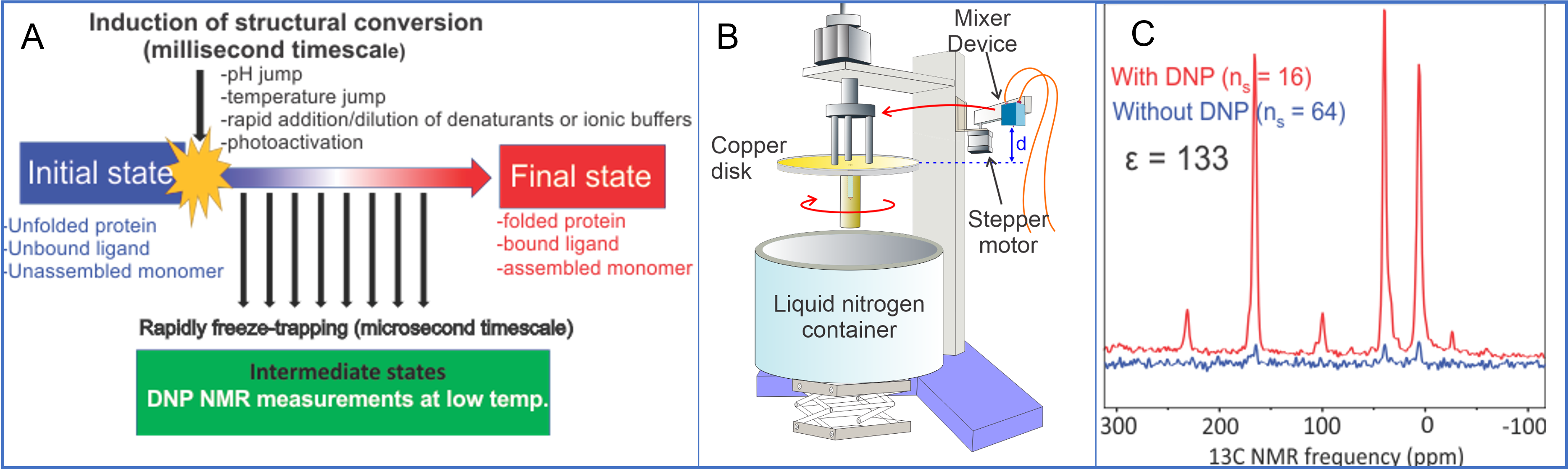
Time resolved solid-state NMR
While dynamics and structural studies on biomolecules at thermal equilibrium conditions by using conventional experimental approaches provide valuable insights into the overall architecture of biomolecules, they often fail to capture the intricate details of their temporal changes and interactions. Especially for kinetically driven reactions, the experimental method with proper time- and spatial resolutions to capture the highly dynamic short-lived intermediate states has been lacking. The recently developed time-resolved solid-state NMR utilizes a rapid-freeze-quenching method and sensitivity-enhanced solid-state NMR via dynamic nuclear polarization (DNP). In our lab, we further develop time-resolved solid-state NMR and apply to clinically and pharmaceutically relevant biomolecules.
Application of Solid-state NMR to assess quality attributes of therapeutics
When assessing the quality attributes of therapeutics, solid-state NMR can provide unique advantages and insights that are not easily obtainable through other methods. In our lab, we aim to develop solid-state NMR measurement techniques for the following therapeutic systems: (1) structural studies on antigen and Aluminum-based adjuvants, and (2) characterization of the high-order structure of monoclonal antibodies.
Protein self-assembly process
A self-assembly process is a universal process ranging from actin/microtubule filament formations in the cell life cycle, viral capsid formations, to pathological fibril formations. However, there has not been an effective experimental approach that can elucidate molecular details as a function of oligomeric size, in particular, at early stages. Recently, we have demonstrated that a combination of two time-resolved approaches, time-resolved ssNMR and light scattering, can characterize molecular structures/arrangements and sizes of oligomers as a function of time. We continue pursuing this approach and elucidate self-assembly processes that can provide potential implications for drug development.
Publications
- Time-resolved solid state NMR of biomolecular processes with millisecond time resolution.
- Nitroxide-based triradical dopants for efficient low-temperature dynamic nuclear polarization in aqueous solutions over a broad pH range.
- Quantitative Agreement between Conformational Substates of Holo Calcium-Loaded Calmodulin Detected by Double Electron-Electron Resonance EPR and Predicted by Molecular Dynamics Simulations.
- Time-resolved DEER EPR and solid-state NMR afford kinetic and structural elucidation of substrate binding to Ca2+-ligated calmodulin.
