Profile
-preview.jpg)
John Orban
Professor
Orban Group (240) 314-6221 jorban@umd.eduDr. Orban’s research interests focus on the area of protein structural biology and design, particularly in understanding how the malleability of protein folds relates to biological function. High field solution NMR spectroscopy and other biophysical and biochemical methods are employed in the laboratory.
CURRENT RESEARCH
Protein switches
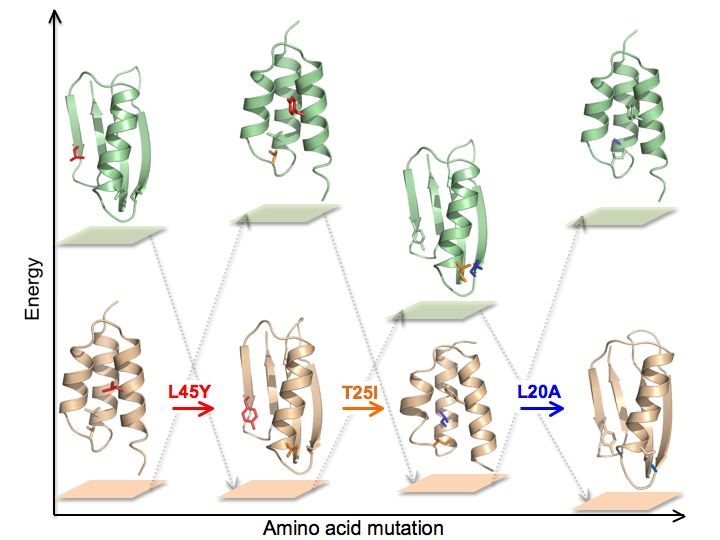
While most globular proteins populate relatively homogeneous conformational ensembles under physiological conditions, significant exceptions continue to emerge. Many biological processes involve extensive re-modeling of protein conformation, including switches from disordered to ordered states. Some natural proteins can even undergo large-scale transitions from one ordered state to another, involving major shifts in secondary structure, repacking of the protein core, and exposure of new surfaces.
Such “metamorphic” proteins are capable of performing alternative functions triggered by binding interactions that stabilize latent conformational states. The ability of these proteins to completely change their fold topologies has implications in a number of important areas including computational and structural biology, protein evolution, human disease, and protein design. The Orban lab is working on the biophysical characterization of protein switches between a number of common fold topologies that occur through short mutational paths or in response to external stimuli.
Intrinsically disordered proteins (IDPs)
The proteins described above are typically on the margin of stability and can be tipped toward one fold or another through relatively subtle changes in sequence or environmental triggers. Intrinsically disordered proteins, on the other hand, have no stable 3D structure and their flexibility allows them to adopt many different conformations. Thus, their structures are characterized by conformational ensembles that are typically non-random coil. These conformational ensembles can be shifted, sometimes dramatically, in response to post-translational modifications or ligand binding. Conceptually, they are similar to metamorphic proteins, having the ability to adopt different structures with different binding partners, for example. The Orban lab is studying an IDP called prostate associated gene 4 (PAGE4) that plays an important role in prostate cancer using a range of biophysical tools including NMR and small angle X-ray scattering (SAXS). PAGE4 undergoes large changes in its conformational ensemble and cellular function depending on the level of phosphorylation.
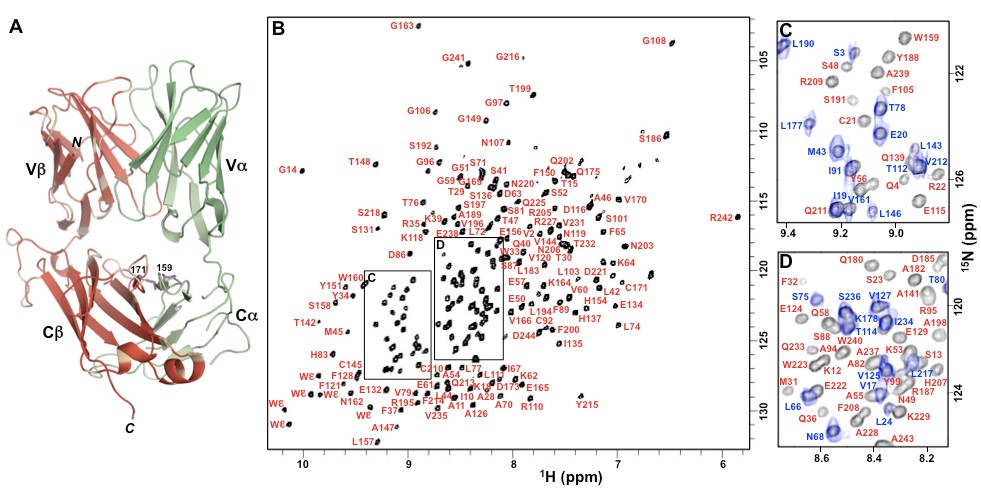
Multi-protein signaling complexes
The conformational changes described above are large amplitude. However, small structural changes can also play an important role in key biological processes. An example of this is
the interaction between peptide-MHC and T cell receptor (TCR) molecules. Although the 3D structures of pMHC, TCR, and pMHC-TCR are known, how this non-covalent binding interaction transmits information to distal TCR-associated CD3 molecules and triggers T cell signaling remains a mystery. The Orban lab is working on characterizing the binding interaction between TCR and CD3 molecules and also understanding how pMHC-binding to TCR leads to allosteric changes that affect the TCR-CD3 interaction.
Publications
- A single C-terminal residue controls SARS-CoV-2 spike trafficking and incorporation into VLPs.
- Acquired resistance to KRAS G12C small-molecule inhibitors via genetic/nongenetic mechanisms in lung cancer.
- Design and characterization of a protein fold switching network.
- Reversible switching between two common protein folds in a designed system using only temperature.
- Structural metamorphism and polymorphism in proteins on the brink of thermodynamic stability.
- Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites.
- Prostate-Associated Gene 4 (PAGE4): Leveraging the Conformational Dynamics of a Dancing Protein Cloud as a Therapeutic Target.
- PAGE4 and Conformational Switching: Insights from Molecular Dynamics Simulations and Implications for Prostate Cancer.
- Phenotypic Plasticity, Bet-Hedging, and Androgen Independence in Prostate Cancer: Role of Non-Genetic Heterogeneity.
- Phosphorylation-induced Conformational Ensemble Switching in an Intrinsically Disordered Cancer/Testis Antigen.
- Identification of the Docking Site for CD3 on the T Cell Receptor β Chain by Solution NMR.
- Subdomain interactions foster the design of two protein pairs with ∼80% sequence identity but different folds.
- Main chain NMR assignments of subtilisin Sbt70 in its prodomain-bound state.
- The design and characterization of two proteins with 88% sequence identity but different structure and function.
- An artificially evolved albumin binding module facilitates chemical shift epitope mapping of GA domain interactions with phylogenetically diverse albumins.
- Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
- Hydrogen-deuterium exchange in free and prodomain-complexed subtilisin.
- Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
- Using offset recombinant polymerase chain reaction to identify functional determinants in a common family of bacterial albumin binding domains.
- Peptidoglycan recognition by Pal, an outer membrane lipoprotein.
- G148-GA3: a streptococcal virulence module with atypical thermodynamics of folding optimally binds human serum albumin at physiological temperatures.
- Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
- Directed evolution of highly homologous proteins with different folds by phage display: implications for the protein folding code.
- HU-alpha binds to the putative double-stranded DNA mimic HI1450 from Haemophilus influenzae.
- NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog.
