Research
Research Interests
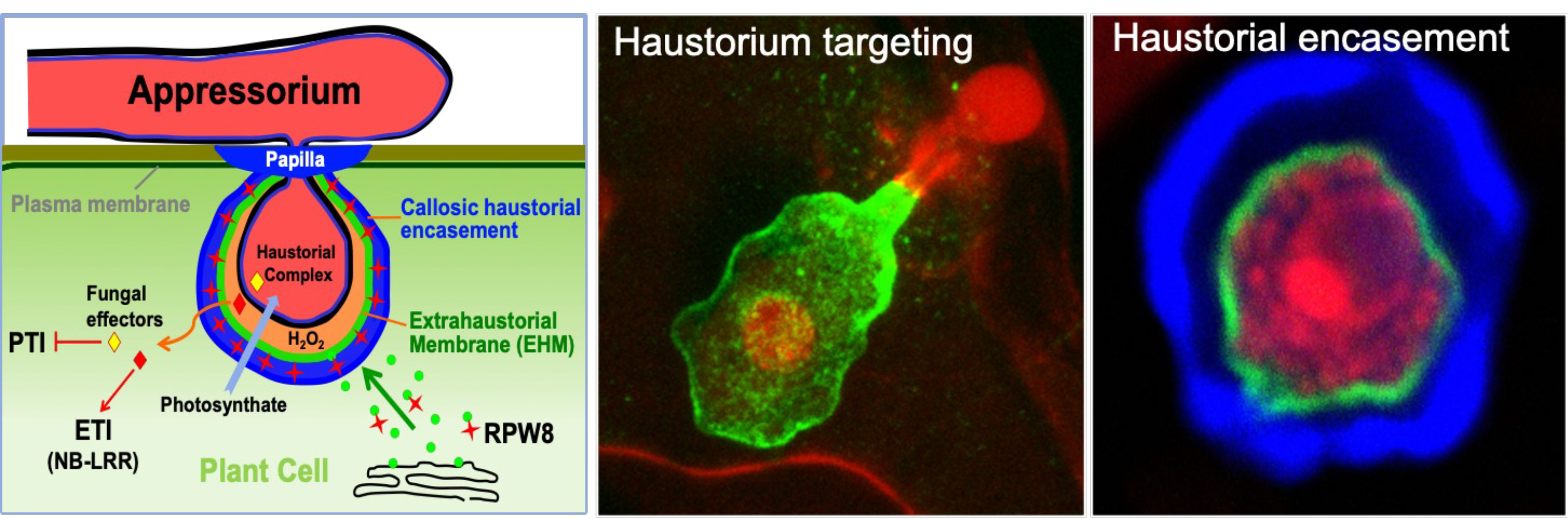
Molecular interaction at the host-pathogen interface:
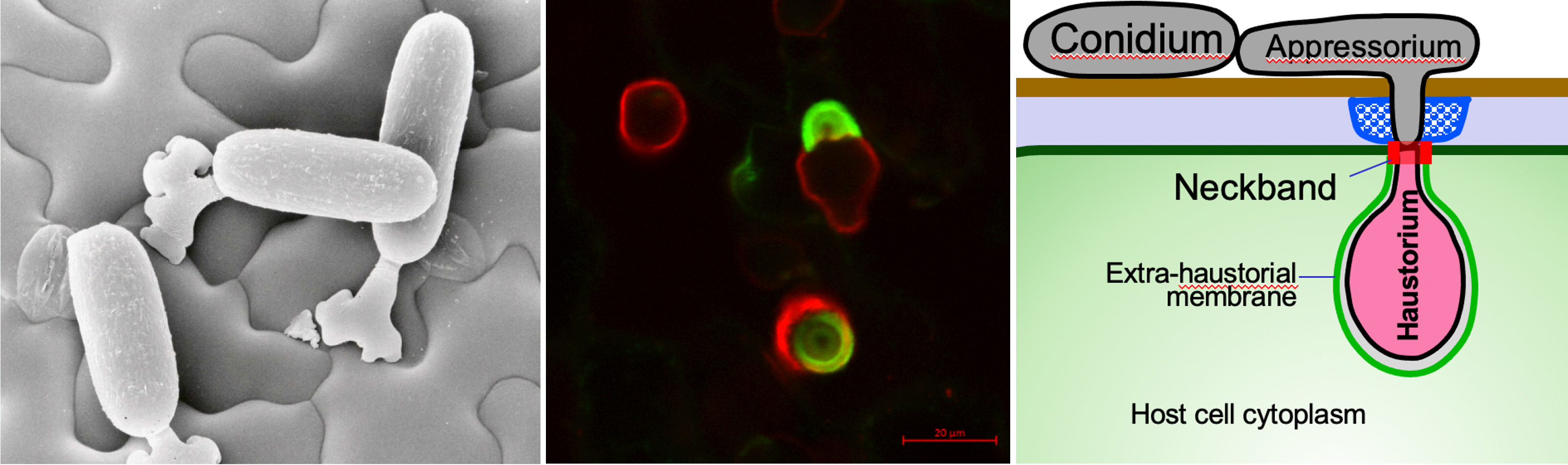
RPW8 is an atypical resistance protein identified in Arabidopsis that is specifically targeted to the extra-haustorial membrane—the enigmatic host-pathogen interface, where RPW8 activates defenses to constrain the fungal feeding structure, i.e. the haustorium. We aim to (i) understand how RPW8 is targeted to, and activates defenses, at the host-pathogen interface, (ii) investigate the the origin & biogenesis of the extra-haustorial membrane and its role in host defense and fungal pathogenesis, (iii) identify more host and fungal proteins interacting at the interfacial membrane, and (iv) engineer novel resistance against powdery mildew in plants.
Dissection of multilayered plant immunity
Plants have evolved effective immune mechanisms to fight off various (potential) pathogens. The complete resistance of all genotypes of a plant species against any non-adapted pathogens is defined as non-host resistance (NHR). We aim to break down NHR of Arabidopsis against distant, non-adapted powdery mildew fungi or even rust fungi to dissect the multilayered plant immunity system through stepwise forward and reverse genetic screens followed by molecular characterization of novel immunity components and their mechanistic connection to well-studies immunity components. A complete dissection of NHR may improve our understanding of the plant immune architecture and inspire novel strategies to boost crop resistance against adapted pathogens. We have engaged a multi-institutional collaboration to explore this line of research. For more information, visit the project website here.

Mechanisms of cell-autonomous plasma membrane repair
Plant cells possess a rigid cell wall, making it difficult to investigate how they detect and respond to plasma membrane injuries at the subcellular level. To colonize their hosts, powdery mildew fungi as biotrophic pathogens must penetrate epidermal cells and establish feeding structures (haustoria) inside living host cells. Because this invasion requires the host plasma membrane to be breached and subsequently maintained, powdery mildew infection serves as an ideal system for studying cell-autonomous plasma membrane repair (CAPMR) in plants. Using forward and reverse genetics alongside cell biological and OMICS approaches, we are identifying and characterizing the genes and proteins that mediate CAPMR. Gaining insight into how plant cells self-repair and how biotrophic fungi manipulate this repair machinery to facilitate infection will inform the development of innovative strategies to combat crop diseases. If you are interested in joining the Xiao Lab to work on this new project? Please email Dr. Xiao at xiao@umd.edu
Pathogenicity mechanisms of powdery mildew fungi
Powdery mildew (PM) is one of the most important and widespread crop diseases that constantly affect plant production. PM is caused by obligate biotrophic ascomycete fungi in the order of Erysiphales. Because PM fungi cannot be cultured or transformed, it is very difficult to study this important group of pathogens by conventional genetic approaches. To tackle this challenge, we have recently sequenced the genomes of four dicot PM biotypes. Through a comprehensive comparative analysis of all available PM genomes, we have identified highly conserved as well as lineage-specific candidate effector genes. Currently, we are conducting comparative genomics-guided, ectopic expression (i.e. in host cells)-enabled, functional studies of key candidate PM genes to understand and intervene PM pathogenesis. We are also utilizing a higher-order Arabidopsis mutant that is super-susceptible to many different non-adapted powdery mildew species for the identification of powdery mildew genes essential for suppressing host immunity.


Current Research in the Xiao Lab is supported by NSF (IOS-2224203), NIH-MIRA (1R35GM158354-01), USDA (NIFA-2022-08786 & 58-8042-5-187) and MAES (MD-PSLA-253688)
