
Osnat Herzberg
Professor
Herzberg Group (240) 314-6245 osnat@umd.eduDr. Osnat Herzberg is a structural biologist interested in the relationship between the function and structure of proteins and how protein-ligand interactions can guide drug discovery. The Herzberg lab uses X-ray crystallography and other biophysical, biochemical, and cellular approaches to better understand various experimental systems.
CURRENT RESEARCH
Current projects in the Herzberg lab include studies of genetic factors that contribute to inflammatory bowel diseases; understanding the structure and function of myovirus phage CBA120 proteins, a bacteriophage that kills pathogenic E. coli; and, drug discovery for combating the parasitic diseases giardiasis and amebiasis.
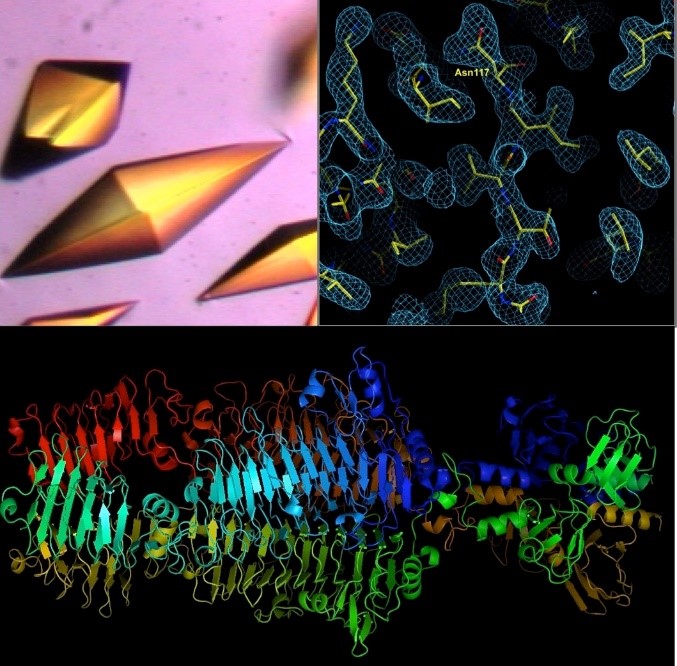
A tailspike protein structure is shown at the bottom.
The inflammatory bowel diseases (IBD), Crohn’s disease and ulcerative colitis, are human complex trait diseases caused by multiple genetic variants and environmental factors. The Herzberg lab is working to identify genetic variants known as single nucleotide polymorphisms (SNPs) that are associated with these diseases. They hope to determine how these variants alter proteins that contribute to disease. This is a collaborative project involving Dr. John Moult’s group. Dr. Moult’s group is identifying candidate disease variants by computational methods and the Herzberg group is carrying out experimental studies to validate these findings. One particular protein, called gasdermin B, has variants that have been implicated in IBD. Studies by the Herzberg lab revealed that protein changes associated with different SNPs alter the flexibility of a protein loop region and these changes have the potential to alter protein-protein interactions. Dr. Herzberg’s lab also discovered that gasdermin B can bind to lipid molecules that are critical to maintaining the integrity of the gut wall. It remains to be determined whether and how the gasdermin B variant found in IBD patients affects the lipid composition of the gut wall.
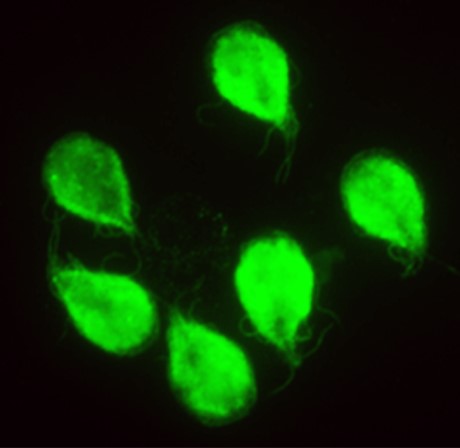
The parasites Giardia lamblia and Entamoeba histolytica cause severe intestinal diseases and are major health hazards in undeveloped and developing countries. Moreover, disease outbreaks in the developed world are associated with drug resistance, which is rapidly spreading, and many patients do not tolerate these standard-of-care drugs. The Herzberg lab is developing less toxic drugs that are not prone to drug resistance. The Herzberg group is optimizing the properties of a drug called fumagillin, which is used in the European Union to treat immune compromised patients suffering from the parasitic infection microsporidiosis, in order to develop new and better antigiardiasis and antiamebiasis therapeutics. Compound optimization is facilitated by the unique capabilities of the group that integrate all aspects of preclinical studies, including medicinal chemistry, biochemistry, molecular biology, cellular biology, and animal studies.
Publications
- Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
- Reply to HU et al.: On the interpretation of gasdermin-B expression quantitative trait loci data.
- A protein-protein interaction dictates Borrelial infectivity.
- Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein.
- Discovery of novel antigiardiasis drug candidates.
- Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
- Crystal structure of ORF210 from E. coli O157:H1 phage CBA120 (TSP1), a putative tailspike protein.
- Structural basis for inactivation of Giardia lamblia carbamate kinase by disulfiram.
- High-throughput Giardia lamblia viability assay using bioluminescent ATP content measurements.
- Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
- X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
- Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
- X-ray structure and kinetic properties of ornithine transcarbamoylase from the human parasite Giardia lamblia.
- Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
- Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
- Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily.
- Swiveling domain mechanism in pyruvate phosphate dikinase.
- Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis.
- Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
- Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
- Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
- Structure and kinetics of phosphonopyruvate hydrolase from Variovorax sp. Pal2: new insight into the divergence of catalysis within the PEP mutase/isocitrate lyase superfamily.
- Crystal structure of the petal death protein from carnation flower.
- Crystal structures representing the Michaelis complex and the thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase.
- Structure of HI0073 from Haemophilus influenzae, the nucleotide-binding domain of a two-protein nucleotidyl transferase.
- Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site.
- Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
- Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation.
- X-ray structure of HI0817 from Haemophilus influenzae: protein of unknown function with a novel fold.
- Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues.
- YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays.
- Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
- Conformational flexibility of PEP mutase.
- Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
- Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot.
- A catalytic mechanism for D-Tyr-tRNATyr deacylase based on the crystal structure of Hemophilus influenzae HI0670.
- Assisting functional assignment for hypothetical Heamophilus influenzae gene products through structural genomics.
- Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR.
- The HI0073/HI0074 protein pair from Haemophilus influenzae is a member of a new nucleotidyltransferase family: structure, sequence analyses, and solution studies.
- Degradation pathway of the phosphonate ciliatine: crystal structure of 2-aminoethylphosphonate transaminase.
- Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae.
- Structure of 2C-methyl-D-erythrol-2,4-cyclodiphosphate synthase from Haemophilus influenzae: activation by conformational transition.
- Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
- From structure to function: YrbI from Haemophilus influenzae (HI1679) is a phosphatase.
- Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis.
- Relocation of the catalytic carboxylate group in class A beta-lactamase: the structure and function of the mutant enzyme Glu166-->Gln:Asn170-->Asp.
- Helix swapping between two alpha/beta barrels: crystal structure of phosphoenolpyruvate mutase with bound Mg(2+)-oxalate.
- Structural consequences of the active site substitution Cys181 ==> Ser in metallo-beta-lactamase from Bacteroides fragilis.
- Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
- Topography of the interaction of HPr(Ser) kinase with HPr.
- A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum.
- Role of the omega-loop in the activity, substrate specificity, and structure of class A beta-lactamase.
- Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
- Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
- Elimination of the hydrolytic water molecule in a class A beta-lactamase mutant: crystal structure and kinetics.
- Structure and kinetics of the beta-lactamase mutants S70A and K73H from Staphylococcus aureus PC1.
- Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis.
- Determination of the nucleotide binding site within Clostridium symbiosum pyruvate phosphate dikinase by photoaffinity labeling, site-directed mutagenesis, and structural analysis.
- Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
- Crystallization of the IIA domain of the glucose permease of Bacillus subtilis.
- Structure of the IIA domain of the glucose permease of Bacillus subtilis at 2.2-A resolution.
- Structural basis for the inactivation of the P54 mutant of beta-lactamase from Staphylococcus aureus PC1.
- Refined crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.0 A resolution.
- Analysis of the steric strain in the polypeptide backbone of protein molecules.
