SMILE and Data Processing
Auto Data Processing Scripts for PCA
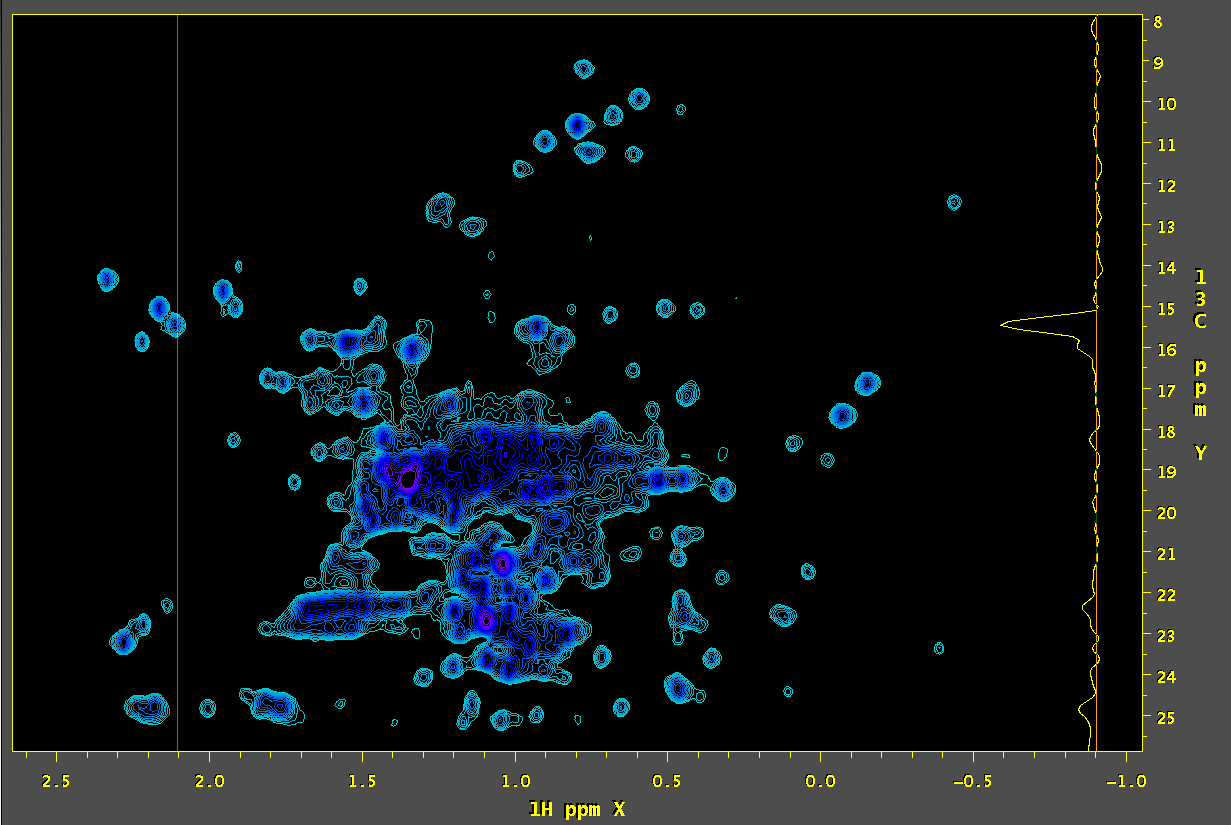
Using SMILE processing to remove residual signals:
For NMRPipe v9.8 and higher, SIERRA-SMILE signal removal can now be accomplished by calling the basicSierra.com function:
basicSierra.com -in test.fid -ft orig.ft2 -out sierra.ft2 \ -reg 2.12 2.07 27.0 25.0 -tdN 6 \ -procArgs -xP0 auto, -xEXT1 3.5ppm -xEXTXN -1.5 ppm \ -xBASEARG POLY,auto ord=4 yP0 -90
where -reg specifies the 1H and 13C ppm bounds for SMILE removal and -tdN specifies the baseline correction order applied to the excipient signal
(To install NMRPipe, go to https://www.ibbr.umd.edu/nmrpipe/install.html.)
An in depth overview of of the SMILE removal process is detailed below:
An explicit processing script can be found here: smile_subtract.txt (rename smile_subtract.com)
1. Process data normally:
#!/bin/csh nmrPipe -in test.fid \ | nmrPipe -fn SOL \ | nmrPipe -fn SP -off 0.50 -end 0.95 -pow 2 -elb 0.0 -glb 0.0 -c 0.5 \ | nmrPipe -fn ZF -zf 1 -auto \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 $ -p1 0.0 -di \ | nmrPipe -fn TP \ | nmrPipe -fn SP -off 0.50 -end 0.95 -pow 1 -elb 0.0 -glb 0.0 -c 0.5 \ | nmrPipe -fn ZF -zf 2 -auto \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 90.0 -p1 0.0 -di \ | nmrPipe -fn TP \ | nmrPipe -fn POLY -auto -ord 0 -verb \ -out orig.ft2 -ov
2. Apply baseline correction in the indirect dimension:
Apply baseline correction in the indirect dimension if needed. As an alternative to the usual method of baseline correction "nmrPipe -fn POLY -auto ...", the "tdBaseline1D.com" utility can baseline correct a spectrum by replacing one or more points at the start of the time-domain data. The "-n ..." parameter specifies the number of points to replace, so that setting "-n 1" will apply a constant baseline correction, and setting higher values will correct for curved baselines.
nmrPipe -in orig.ft2 -fn TP -out tmp.ft2 -ov tdBaseline1D.com -in tmp.ft2 -out base.ft2 -n $ /bin/rm -f tmp.ft2
3. Reprocess the baseline corrected data with SMILE (set -report 2 to save SMILE peak table):
nmrPipe -in base.ft2 \ | nmrPipe -fn HT \ | nmrPipe -fn PS -inv -hdr \ | nmrPipe -fn FT -inv \ | nmrPipe -fn ZF -inv \ | nmrPipe -fn SP -inv -hdr \ | nusPipe -fn SMILE -nDim 2 -maxIter 2048 -nSigma 2 -report 2 -sample None \ -xApod SP -xQ1 0.50 -xQ2 0.95 -xQ3 1 -xELB 0.0 -xGLB 0.0 \ -xT 100 -xP0 -90.0 -xP1 0.0 -scaling 1.0 \ | nmrPipe -fn ZF -zf 1 -auto \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 -100.0 -p1 0.0 -di \ | nmrPipe -fn TP \ -out smile.ft2 -ov nmrPipe -in base.ft2 -out base.ft2 -inPlace -fn TP
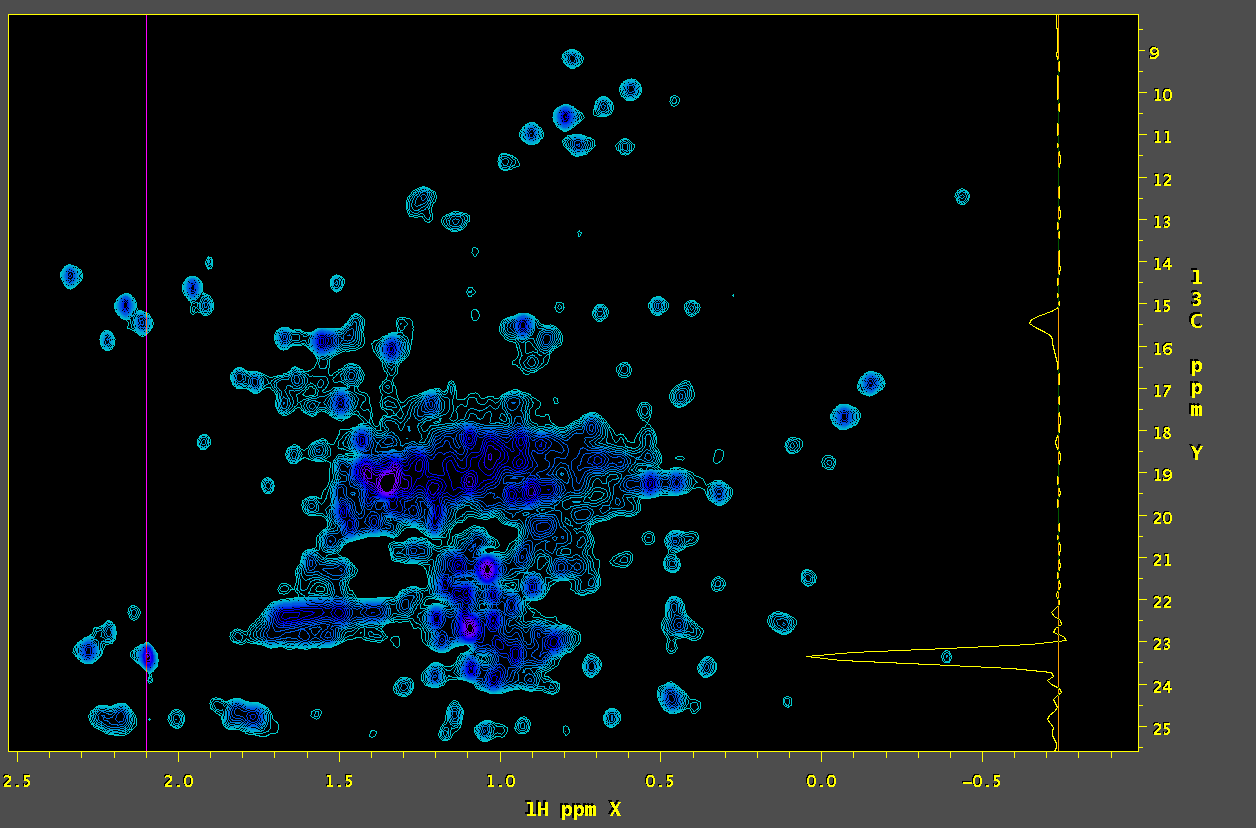
4. Extract the relevant peaks from the SMILE peak table by setting the bounds around the peak of interest including attendant artifacts:
smile2tab.tcl -in smile.log -out smile.tab -ref orig.ft2 selectTab.tcl -in smile.tab -out small.tab -hdr -cond "X_PPM <= $downfield-1H && X_PPM >= $upfield-1H && Y_PPM <= $downfield-13C && Y_PPM >= $upfield-13C"
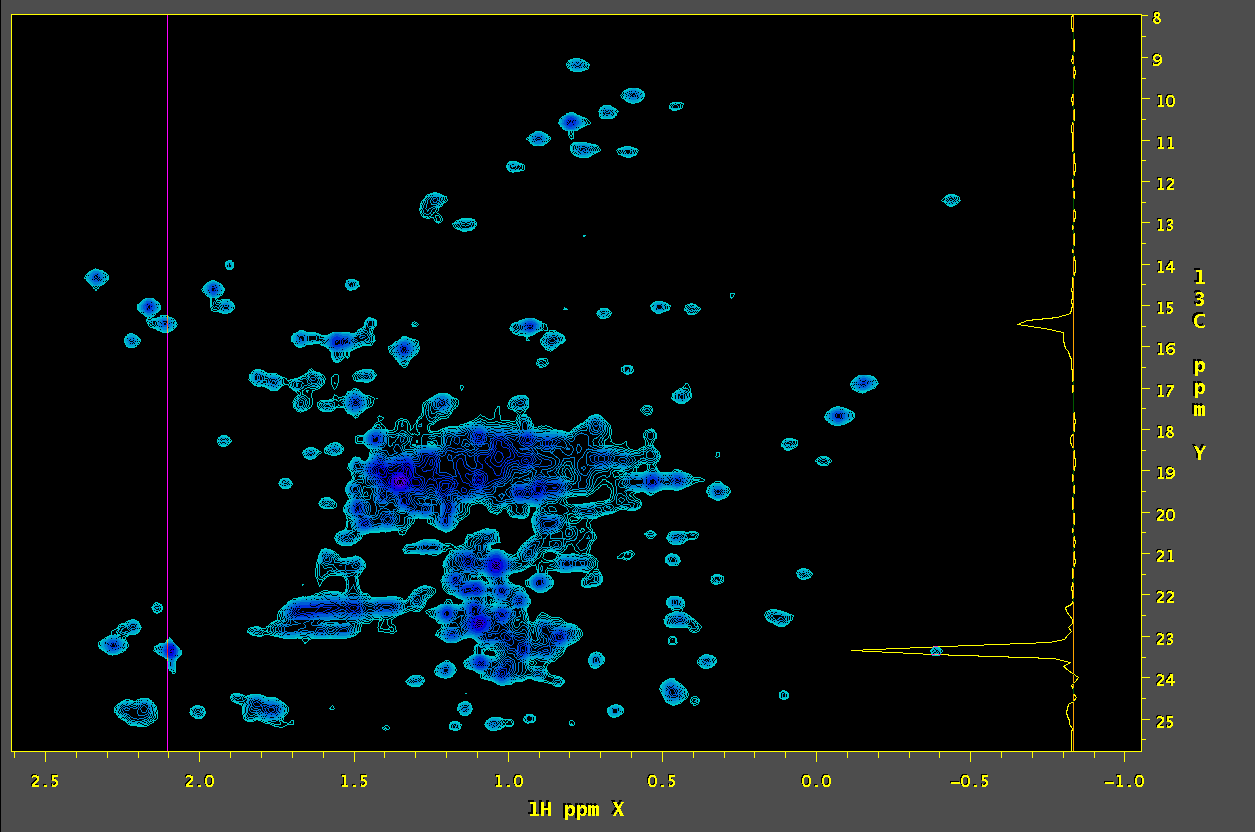
5. Process the simulated time-domain SMILE data:
set xT = (`getParm -in orig.ft2 -parm NDTDSIZE -fmt %.0f -dim CUR_XDIM`) set yT = (`getParm -in orig.ft2 -parm NDTDSIZE -fmt %.0f -dim CUR_YDIM`) set xN = (`getParm -in smile.ft2 -parm NDFTSIZE -fmt %.0f -dim CUR_XDIM`) set yN = (`getParm -in smile.ft2 -parm NDFTSIZE -fmt %.0f -dim CUR_YDIM`) set xP0 = (`getParm -in orig.ft2 -parm NDP0 -fmt %.1f -dim CUR_XDIM`) set yP0 = (`getParm -in orig.ft2 -parm NDP0 -fmt %.1f -dim CUR_YDIM`) echo "" echo "SMILE Simulation: Time-Domain Size: $xT $yT Freq-Domain Size: $xN $yN" simTimeND -in small.tab -tdd -nots -xP0 $xP0 -yP0 $yP0 \ -xN $xN -yN $yN \ -xT $xT -yT $yT \ -xMODE DQD -yMODE Echo-AntiEcho \ -xSW 12626.263 -ySW 5434.783 \ -xOBS 900.162 -yOBS 226.349 \ -xCAR 4.727 -yCAR 18.000 \ -xLAB 1H -yLAB 13C \ -ndim 2 -aq2D Complex \ -out ./sim.fid -verb -ov nmrPipe -in sim.fid \ | nmrPipe -fn SP -off 0.50 -end 0.95 -pow 2 -elb 10.0 -glb 0.0 -c 0.5 \ | nmrPipe -fn ZF -zf 1 -auto \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 $xP0 -p1 0.0 -di \ | nmrPipe -fn TP \ | nmrPipe -fn SP -off 0.50 -end 0.95 -pow 1 -elb 0.0 -glb 0.0 -c 0.5 \ | nmrPipe -fn ZF -zf 2 -auto \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 $yP0 -p1 0.0 -di \ | nmrPipe -fn TP \ -out sim.ft2 -ov
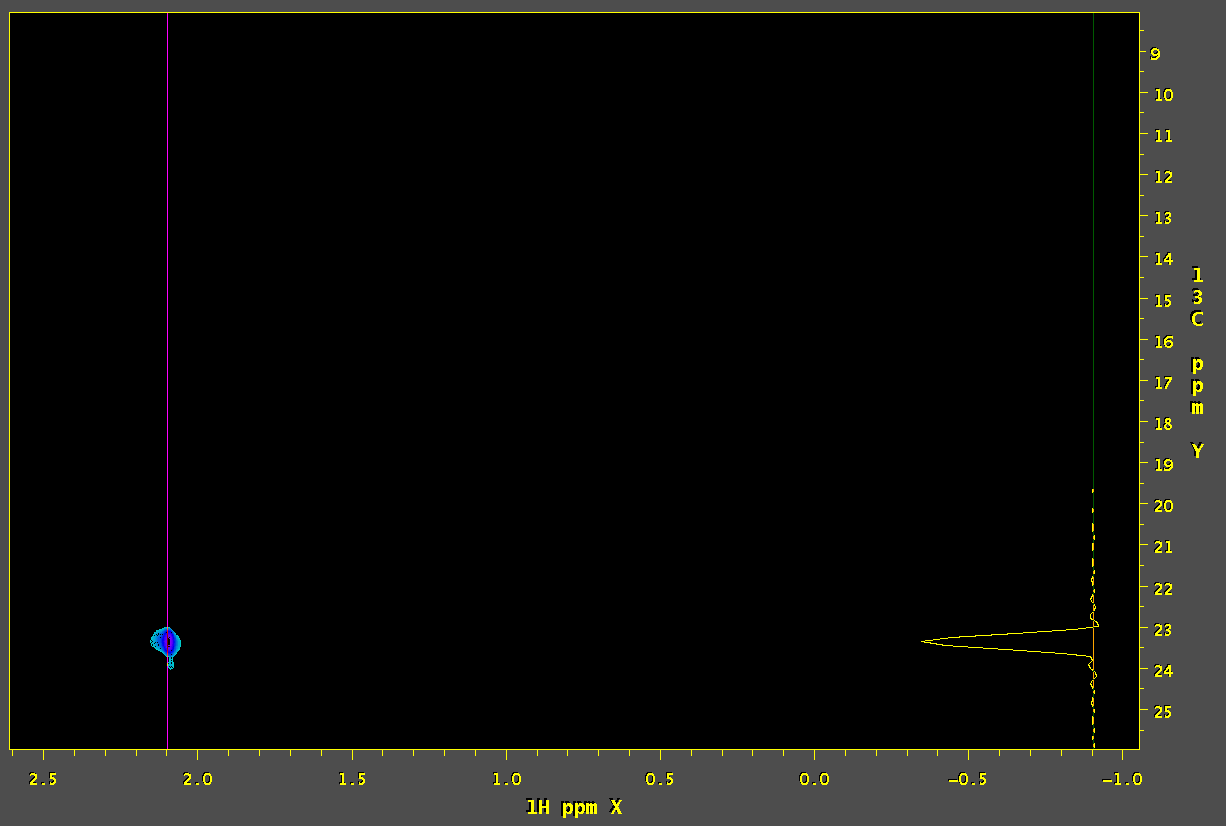
6. Scale simulated SMILE data using bounds set tightly around peak of interest and subtract from original measured data:
set c = (`findScale.tcl -in1 orig.ft2 -in2 sim.ft2 -roiArg -x X_AXIS $1Hppm $1Hppm -y Y_AXIS $13Cppm $13Cppm`) echo $c nmrPipe -in sim.ft2 -out sim.ft2 -inPlace -fn MULT -c $c addNMR -in1 orig.ft2 -in2 sim.ft2 -out dif.ft2 -sub sethdr orig.ft2 -title Measured sethdr base.ft2 -title Baseline sethdr smile.ft2 -title SMILE sethdr sim.ft2 -title Simulated sethdr dif.ft2 -title Difference scale2D *.ft2